| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
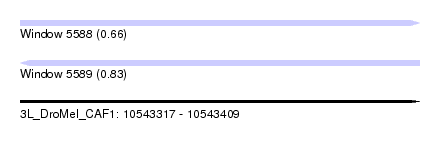
| Location | 10,543,317 – 10,543,409 |
| Length | 92 |
| Max. P | 0.832831 |

| Location | 10,543,317 – 10,543,409 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -22.49 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
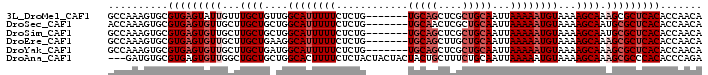
>3L_DroMel_CAF1 10543317 92 + 23771897 UGUUGGUGUGAGCGCUUUGCUUUUACAUUUUUAAUUGCAGCGAGCUGCA-------CAGAGAAAAAUGCCAACAGCAAACAAUACUCACGCACUUUGGC .....(((((((...((((((.((.(((((((...(((((....)))))-------.....)))))))..)).)))))).....)))))))........ ( -26.20) >DroSec_CAF1 422756 92 + 1 UGUUGGUGUGAGCGCAUUGCUUUUACAUUUUUAAUUGCAGCGAGUUGCA-------CAGAGAAAAAUGCCAGCAGCAAGCAACACUCACGCACUUUGGU .....(((((((.((.(((((.((.(((((((...(((((....)))))-------.....)))))))..)).)))))))....)))))))........ ( -25.00) >DroSim_CAF1 431237 92 + 1 UGUUGGUGUGAGCGCAUUGCUUUUACAUUUUUAAUUGCAGCGAGCUGCA-------CAGAGAAAAAUGCCAGCAGCAAGCAACACUCACGCACUUUGGC .....(((((((.((.(((((.((.(((((((...(((((....)))))-------.....)))))))..)).)))))))....)))))))........ ( -27.40) >DroEre_CAF1 431632 92 + 1 UGUUGGUGUGAGCGCUUUGCUUUUACAUUUUUAAUUGCAGCAAGCUGCA-------CAGAGAAAAAUGCCUUCAGCAAGCAACACUCACGCACUUUGGC .....(((((((.((((.(((....(((((((...(((((....)))))-------.....))))))).....)))))))....)))))))........ ( -27.40) >DroYak_CAF1 431938 92 + 1 UGUUGGUGUGAGCGCUUUGCUUUUACAUUUUUAAUUGCAGCGAGCUGCA-------CAGAGAAAAAUGCCAUCAGCAAGCAACACUCACGCACUUUGGC .....(((((((.((((.(((....(((((((...(((((....)))))-------.....))))))).....)))))))....)))))))........ ( -27.40) >DroAna_CAF1 416648 96 + 1 UCUGGGUGUGGGCGCUUUGCUUUUACAUUUUUAAUUGCAGAAAGCAGUAGUAGUAGUAGAGAAAAGUGCCAGCAGCAGCCAACACUCACGCACAUC--- ..(((((((.((((((((..(((((((((....(((((.....)))))...))).)))))).)))))))).((....))..)))))))........--- ( -28.90) >consensus UGUUGGUGUGAGCGCUUUGCUUUUACAUUUUUAAUUGCAGCGAGCUGCA_______CAGAGAAAAAUGCCAGCAGCAAGCAACACUCACGCACUUUGGC .....(((((((.(..(((((....(((((((...(((((....)))))............))))))).....)))))....).)))))))........ (-22.49 = -21.83 + -0.66)

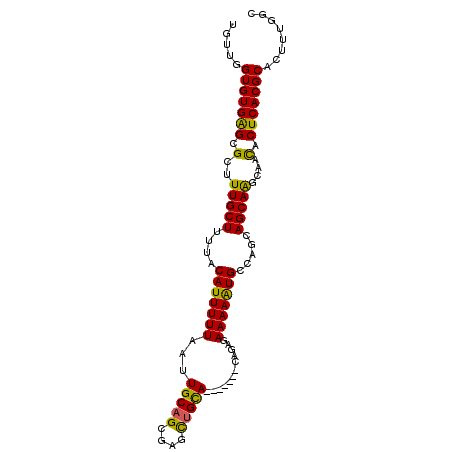
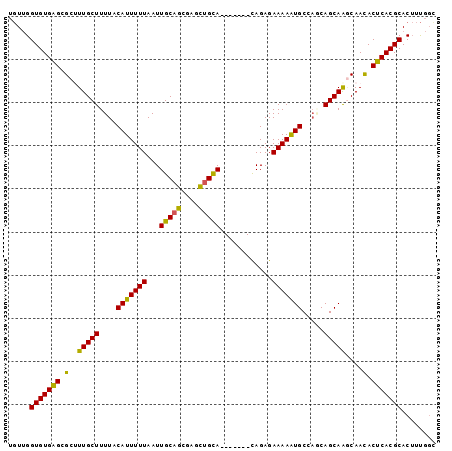
| Location | 10,543,317 – 10,543,409 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -23.53 |
| Energy contribution | -24.86 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10543317 92 - 23771897 GCCAAAGUGCGUGAGUAUUGUUUGCUGUUGGCAUUUUUCUCUG-------UGCAGCUCGCUGCAAUUAAAAAUGUAAAAGCAAAGCGCUCACACCAACA ((......))((((((....((((((....((((((((.....-------(((((....)))))...))))))))...))))))..))))))....... ( -29.10) >DroSec_CAF1 422756 92 - 1 ACCAAAGUGCGUGAGUGUUGCUUGCUGCUGGCAUUUUUCUCUG-------UGCAACUCGCUGCAAUUAAAAAUGUAAAAGCAAUGCGCUCACACCAACA ..........((((((((...(((((....((((((((.....-------((((......))))...))))))))...))))).))))))))....... ( -28.80) >DroSim_CAF1 431237 92 - 1 GCCAAAGUGCGUGAGUGUUGCUUGCUGCUGGCAUUUUUCUCUG-------UGCAGCUCGCUGCAAUUAAAAAUGUAAAAGCAAUGCGCUCACACCAACA ((......))((((((((...(((((....((((((((.....-------(((((....)))))...))))))))...))))).))))))))....... ( -33.10) >DroEre_CAF1 431632 92 - 1 GCCAAAGUGCGUGAGUGUUGCUUGCUGAAGGCAUUUUUCUCUG-------UGCAGCUUGCUGCAAUUAAAAAUGUAAAAGCAAAGCGCUCACACCAACA ((......))(((((((((..(((((....((((((((.....-------(((((....)))))...))))))))...))))))))))))))....... ( -33.30) >DroYak_CAF1 431938 92 - 1 GCCAAAGUGCGUGAGUGUUGCUUGCUGAUGGCAUUUUUCUCUG-------UGCAGCUCGCUGCAAUUAAAAAUGUAAAAGCAAAGCGCUCACACCAACA ((......))(((((((((..(((((....((((((((.....-------(((((....)))))...))))))))...))))))))))))))....... ( -33.30) >DroAna_CAF1 416648 96 - 1 ---GAUGUGCGUGAGUGUUGGCUGCUGCUGGCACUUUUCUCUACUACUACUACUGCUUUCUGCAAUUAAAAAUGUAAAAGCAAAGCGCCCACACCCAGA ---..((.((((((((((..((....))..)))))).................((((((.((((........))))))))))..)))).))........ ( -26.60) >consensus GCCAAAGUGCGUGAGUGUUGCUUGCUGCUGGCAUUUUUCUCUG_______UGCAGCUCGCUGCAAUUAAAAAUGUAAAAGCAAAGCGCUCACACCAACA ..........(((((((((...((((....((((((((............(((((....)))))...))))))))...)))).)))))))))....... (-23.53 = -24.86 + 1.33)

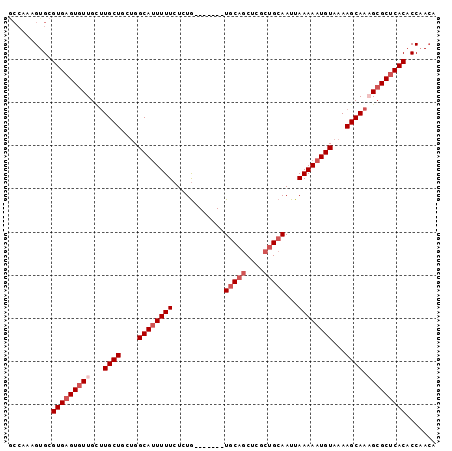
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:34 2006