
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,542,044 – 10,542,187 |
| Length | 143 |
| Max. P | 0.749942 |

| Location | 10,542,044 – 10,542,155 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -27.21 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
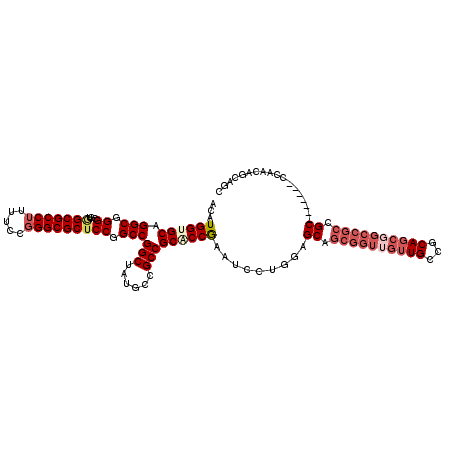
>3L_DroMel_CAF1 10542044 111 + 23771897 U-CCGUUUCUUUUUUCUC-UUUU--GAACAUGCAGCACAUGGUGCAGGCGGGCGUAGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGU .-(((((.(((..(((..-....--)))....(((....((((((.(((((.((.(((((((.....))))))).)).))(((...))))))))))))....)))))).))))). ( -48.00) >DroVir_CAF1 469219 106 + 1 ---CCUUUGU-----CUGCCUCU-CUCAUUUGCAGCACAUGGUGCAGGCUGGCGUGGCGCCUUUUCCAGGCGCACCAGCUGGCUAUGCCGCAGCACCUGGACCGGCCGCUGCAGC ---.......-----........-.....(((((((....(((((.((((((....((((((.....)))))).))))))(((...)))...))))).((.....))))))))). ( -44.60) >DroPse_CAF1 454141 107 + 1 U-CCGUCUCUCUUU-----AUCU--AUCCAUACAGCACAUGGUGCAGGCUGGCGUGGCGCCAUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCUCCAGGGCAGGCAGCAGCGGC .-((((........-----....--.........((((...)))).((((((((..(((((.......)))))..).))))))).(((.((((((....))).))).))))))). ( -44.50) >DroGri_CAF1 432488 103 + 1 ---CUUUUCC-----CCG-UUCUCAUCAUUUACAGCACAUGGUGCAGACUGGAGUG---CCGUUUUCGGGCGCCCCGGCUGCCUAUGCGGCCGCACCAGGACCGGCGGCUGCGGC ---.......-----((.-...................((((.((((.((((.(((---((.......))))).))))))))))))((((((((..........)))))))))). ( -41.60) >DroSim_CAF1 429788 111 + 1 U-CCGUUUCUUUUUUCUC-UUCU--GAACAUGCAGCACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGU .-(((((.((((.(((..-..((--(......))).....(((((.(((((.((..((((((.....))))))..)).))(((...))))))))))))))....)))).))))). ( -48.70) >DroAna_CAF1 415566 106 + 1 UGACAUUUUUUUUGU-------C--CAAAAUGCAGCACAUGGUGCAGGCGGGCGUAGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCUCCAGGUCCGGCAGCAGCAGC .((((.......)))-------)--.....(((.((....(((..((.(((.((.(((((((.....))))))).)).))).))..)))((((.........)))).)).))).. ( -44.60) >consensus U_CCGUUUCUUUUU_CU__UUCU__GAACAUGCAGCACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCAGGUCCGGCAGCAGCGGC ..............................(((.((.(.((((((.(((.(((((((((((.......)))((....))..)))))))))))))))))......)..)).))).. (-27.21 = -28.60 + 1.39)


| Location | 10,542,044 – 10,542,155 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.94 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -27.16 |
| Energy contribution | -28.13 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10542044 111 - 23771897 ACCGCUGCUCCAGGAUUCGGUGCGGCGGCAUAGCCGGCCGGAGCGCCCGGAAAAGGCGCUACGCCCGCCUGCACCAUGUGCUGCAUGUUC--AAAA-GAGAAAAAAGAAACGG-A .(((...((.(((.((..(((((((((((...)))((.((.((((((.......)))))).)).))))).)))))..)).)))....(((--....-..)))...))...)))-. ( -42.10) >DroVir_CAF1 469219 106 - 1 GCUGCAGCGGCCGGUCCAGGUGCUGCGGCAUAGCCAGCUGGUGCGCCUGGAAAAGGCGCCACGCCAGCCUGCACCAUGUGCUGCAAAUGAG-AGAGGCAG-----ACAAAGG--- (((((((((.((......)))))))))))...((..((((((((((((.....))))))...))))))..)).((...((((((...(...-.)..))))-----.))..))--- ( -51.00) >DroPse_CAF1 454141 107 - 1 GCCGCUGCUGCCUGCCCUGGAGCGGCGGCAUAGCCGGCCGGAGCGCCCGGAAAUGGCGCCACGCCAGCCUGCACCAUGUGCUGUAUGGAU--AGAU-----AAAGAGAGACGG-A (((((((((.((......)))))))))))...((.(((.((.(((((.......)))))....)).))).)).(((((.....)))))..--....-----............-. ( -45.30) >DroGri_CAF1 432488 103 - 1 GCCGCAGCCGCCGGUCCUGGUGCGGCCGCAUAGGCAGCCGGGGCGCCCGAAAACGG---CACUCCAGUCUGCACCAUGUGCUGUAAAUGAUGAGAA-CGG-----GGAAAAG--- ((((....((...((((((((...(((.....))).))))))))...))....)))---).((((..(((.((.(((.........))).))))).-.))-----)).....--- ( -37.30) >DroSim_CAF1 429788 111 - 1 ACCGCUGCUCCAGGAUUCGGUGCGGCGGCAUAGCCGGCCGGAGCGCCCGGAAAAGGCGCCACGCCCGCCUGCACCAUGUGCUGCAUGUUC--AGAA-GAGAAAAAAGAAACGG-A .(((...((.(((.((..(((((((((((...)))((.((..(((((.......)))))..)).))))).)))))..)).)))....(((--....-..)))...))...)))-. ( -41.80) >DroAna_CAF1 415566 106 - 1 GCUGCUGCUGCCGGACCUGGAGCGGCGGCAUAGCCGGCCGGAGCGCCCGGAAAAGGCGCUACGCCCGCCUGCACCAUGUGCUGCAUUUUG--G-------ACAAAAAAAAUGUCA (((((((((.(((....))))))))))))...((.(((...((((((.......))))))..))).))..((((...)))).(((((((.--.-------......))))))).. ( -45.50) >consensus GCCGCUGCUGCCGGACCUGGUGCGGCGGCAUAGCCGGCCGGAGCGCCCGGAAAAGGCGCCACGCCAGCCUGCACCAUGUGCUGCAUGUUA__AGAA__AG_AAAAAGAAACGG_A ..((((.(..........).))))((((((((((.(((.((.(((((.......)))))....)).))).))....))))))))............................... (-27.16 = -28.13 + 0.98)



| Location | 10,542,076 – 10,542,187 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -60.52 |
| Consensus MFE | -44.42 |
| Energy contribution | -46.33 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10542076 111 + 23771897 ACAUGGUGCAGGCGGGCGUAGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCGGCCGCCGC-------CCAACAACAGC ...((((((.(((((.((.(((((((.....))))))).)).))(((...)))))))))))).....(((.((.((((((((((...)))))))))).))-------)))........ ( -59.90) >DroSec_CAF1 421545 111 + 1 ACAUGGUGCAGGCGGGCGUGGCGCCUUUCCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCGGCCGCCGC-------CCAACAGCAGC ...((((((.(((((.((..((((((.....))))))..)).))(((...)))))))))))).....(((.((.((((((((((...)))))))))).))-------)))........ ( -60.50) >DroSim_CAF1 429820 111 + 1 ACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCGGCCGCCGC-------CCAACAACAGC ...((((((.(((((.((..((((((.....))))))..)).))(((...)))))))))))).....(((.((.((((((((((...)))))))))).))-------)))........ ( -60.50) >DroEre_CAF1 430475 111 + 1 ACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCUCCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCGGCCGCCGC-------CCAACAGCAGC ...((.((..(((((.((..((((((.....))))))..)).))(((...(((((.((((((.....)))))).(((((....))))).))))).)))))-------)...)).)).. ( -61.20) >DroYak_CAF1 430761 111 + 1 ACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCUCCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCCGCCGCCGC-------CCAACAGCAGC ...(((.((.(((((.((..((((((.....))))))..)(((((((...(((((.((((((.....)))))).)))))....))))..)))))))))))-------)))........ ( -61.30) >DroMoj_CAF1 539142 100 + 1 ACAUGGUGCAGGCUGGCGUGGCGCCGUUUCCAGGCGCACCAGCCGGCUAUCCUGCCGCGCCAGGACCAGCGGCAGCGGCCGC---------------UGCUGGUGCGCCCACAGC--- ...(((.((.((((((....(((((.......))))).))))))(((......)))(((((((...(((((((....)))))---------------))))))))))))))....--- ( -59.70) >consensus ACAUGGUGCAGGCGGGCGUGGCGCCUUUUCCGGGCGCUCCGGCCGGCUAUGCCGCCGCACCGAAUCCUGGAGCAGCGGUUGUUGCCGCAGCGGCCGCCGC_______CCAACAGCAGC ...((((((.(((.((...(((((((.....))))))))).)))(((......))))))))).........((.((((((((((...)))))))))).)).................. (-44.42 = -46.33 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:31 2006