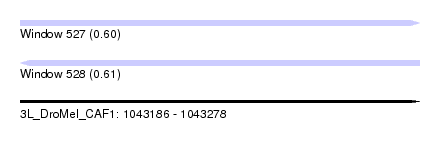
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,043,186 – 1,043,278 |
| Length | 92 |
| Max. P | 0.605407 |

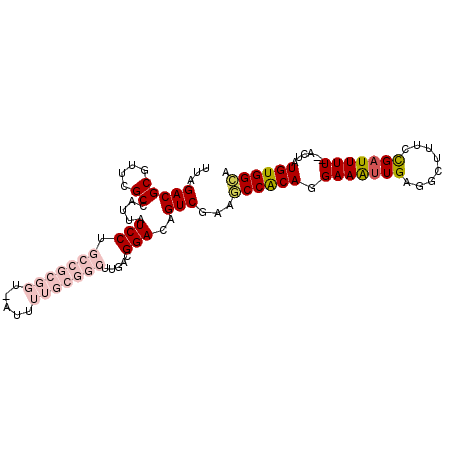
| Location | 1,043,186 – 1,043,278 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1043186 92 + 23771897 UGCCACAUAGU--AAAAUCGGAAAGCCUCAAUUUCCUGUGGCUACGACUGUCCGUCAAGCCGCAAAAUGACCGCGGCAGGAUAAUGG------CGUCUAA .((((......--....(((...((((.((......)).)))).))).(((((.....(((((.........))))).))))).)))------)...... ( -28.50) >DroSec_CAF1 17859 88 + 1 UGCCACAUAGUCGAAAAUCGGAAAGCCUCAAUUUCCUGUGGCUUCGACUGUCCGCCAA------------ACGCGGCAGGAUAAUGGCGAACGCGUCUAA .((((...(((((((.((.(((((.......))))).))...)))))))(((((((..------------....))).))))..))))............ ( -27.90) >DroSim_CAF1 18620 88 + 1 UGCCACAUAGUCGAAAAUCGGAAAGCCUCAAUUUCCUGUGGCUUCGACUGUCCGCCAA------------CCGCGGCAGGAUAAUGGCAAACGCGUCUAA (((((...(((((((.((.(((((.......))))).))...)))))))(((((((..------------....))).))))..)))))........... ( -28.60) >DroEre_CAF1 20905 98 + 1 UACCACAUGGU--AAAAGCAGAAAGCCCCAACUUCCUGCGGUUUCGACUGUCCGUAAAGCCGCAAAAUAACCGCGGCAGGAUAAUGGCGAACGCGUCUAA ((((....)))--)...((((.(((......))).))))......((((((((.....(((((.........))))).)))))...((....)))))... ( -28.10) >DroYak_CAF1 18591 98 + 1 UGCCACAUGGU--AAAAUCGGAAAGCCCCAAUUUCCUGCGGUUUUGACUGUCCGUCAAGCCGCAAAAUAACCGCGGCAGGAUAAUGGCGAACGCGUCUAA ((((...((((--......(((((.......)))))((((((((.(((.....))))))))))).....)))).)))).......((((....))))... ( -30.30) >consensus UGCCACAUAGU__AAAAUCGGAAAGCCUCAAUUUCCUGUGGCUUCGACUGUCCGUCAAGCCGCAAAAU_ACCGCGGCAGGAUAAUGGCGAACGCGUCUAA (((((............(((..(((((.((......)).))))))))(((.((((.................))))))).....)))))........... (-17.35 = -17.83 + 0.48)

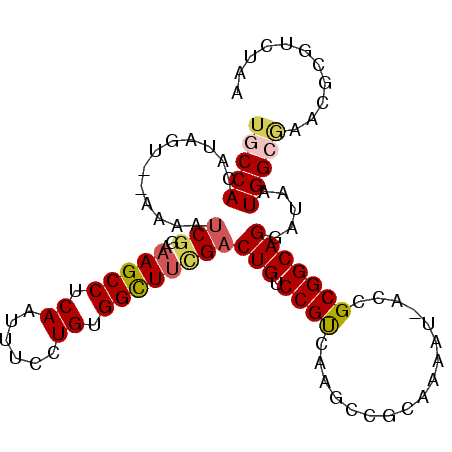
| Location | 1,043,186 – 1,043,278 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -17.73 |
| Energy contribution | -20.18 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1043186 92 - 23771897 UUAGACG------CCAUUAUCCUGCCGCGGUCAUUUUGCGGCUUGACGGACAGUCGUAGCCACAGGAAAUUGAGGCUUUCCGAUUUU--ACUAUGUGGCA ...(((.------((.(((....(((((((.....))))))).))).))...)))...((((((.(((((((........)))))))--....)))))). ( -33.10) >DroSec_CAF1 17859 88 - 1 UUAGACGCGUUCGCCAUUAUCCUGCCGCGU------------UUGGCGGACAGUCGAAGCCACAGGAAAUUGAGGCUUUCCGAUUUUCGACUAUGUGGCA ...(((..((((((((..............------------.)))))))).)))...((((((((((((((........)))))))).....)))))). ( -31.06) >DroSim_CAF1 18620 88 - 1 UUAGACGCGUUUGCCAUUAUCCUGCCGCGG------------UUGGCGGACAGUCGAAGCCACAGGAAAUUGAGGCUUUCCGAUUUUCGACUAUGUGGCA ...(((..((((((((....((......))------------.)))))))).)))...((((((((((((((........)))))))).....)))))). ( -31.60) >DroEre_CAF1 20905 98 - 1 UUAGACGCGUUCGCCAUUAUCCUGCCGCGGUUAUUUUGCGGCUUUACGGACAGUCGAAACCGCAGGAAGUUGGGGCUUUCUGCUUUU--ACCAUGUGGUA ...(((..(((((....((....(((((((.....)))))))..))))))).)))......(((((((((....)))))))))...(--(((....)))) ( -33.20) >DroYak_CAF1 18591 98 - 1 UUAGACGCGUUCGCCAUUAUCCUGCCGCGGUUAUUUUGCGGCUUGACGGACAGUCAAAACCGCAGGAAAUUGGGGCUUUCCGAUUUU--ACCAUGUGGCA ...(((..(((((.((.......(((((((.....))))))).)).))))).)))....(((((.(((((((((....)))))))))--....))))).. ( -34.10) >consensus UUAGACGCGUUCGCCAUUAUCCUGCCGCGGU_AUUUUGCGGCUUGACGGACAGUCGAAGCCACAGGAAAUUGAGGCUUUCCGAUUUU__ACUAUGUGGCA ...(((((....)).....(((.(((((((.....))))))).....)))..)))...((((((.(((((((........)))))))......)))))). (-17.73 = -20.18 + 2.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:59 2006