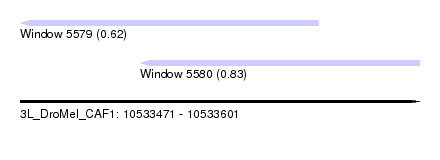
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,533,471 – 10,533,601 |
| Length | 130 |
| Max. P | 0.834498 |

| Location | 10,533,471 – 10,533,568 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10533471 97 - 23771897 UGGCUCUGGCCACU-------GGUCACUUGGCCACUGGACUACAUAGCAUUGGAUACUACCAACCAAACCCACCACCUGCUGGUUGGUCAUUUAGCCUAAGCGC ..(((..(((((.(-------((((....))))).))(((((..(((((((((......))))..............)))))..))))).....)))..))).. ( -28.73) >DroSec_CAF1 413074 102 - 1 UGGCUCUGGCCACUG--CAGUGGUCACUUGGCCACUGGACUACAUAGCACUGGAUACUCGCAACCAAACCCACCACCUGCUGAUUGGUCAUUUAGCCUAAGCGC ..(((..(((.....--((((((((....))))))))(((((..(((((.(((...................)))..)))))..))))).....)))..))).. ( -33.31) >DroSim_CAF1 421460 97 - 1 UGGCUCUGGCCACU-------GGUCACUUGGCCACUGGACUCCAUAGCACUGGAUACUAACAACCAAACCCACCACCUGCUGGUUGGUCAUUUAGCCUAAGCGC ..(((..(((((.(-------((((....))))).))(((((((......))))......((((((..............))))))))).....)))..))).. ( -29.04) >DroYak_CAF1 422219 96 - 1 UGGUACUGGCCACUGGCCACUGGUCACUCGGCCACUGGACAACAUAGCACUAGAUACUCC-----A---CCACCACCUGCUGGUUGGUCAUUUAGCCUAAGCGC ((((..(((((..(((((...)))))...))))).((((.....((....)).....)))-----)---..))))...(((((((((....))))))..))).. ( -25.70) >consensus UGGCUCUGGCCACU_______GGUCACUUGGCCACUGGACUACAUAGCACUGGAUACUACCAACCAAACCCACCACCUGCUGGUUGGUCAUUUAGCCUAAGCGC ..(((..(((...........((((....))))....((((...(((((.(((...................)))..)))))...)))).....)))..))).. (-20.90 = -21.46 + 0.56)

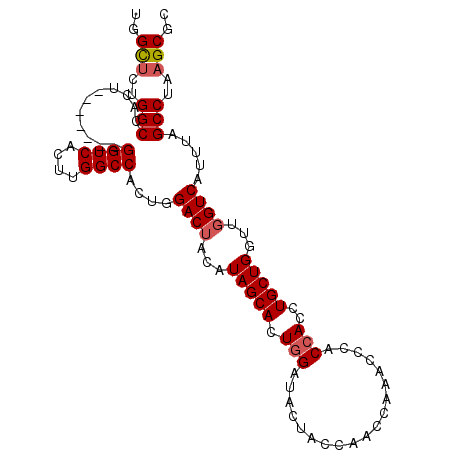
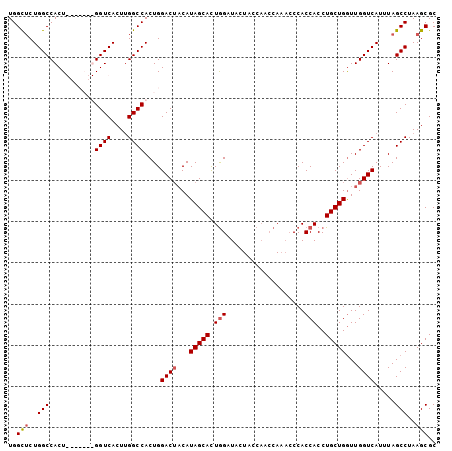
| Location | 10,533,510 – 10,533,601 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -21.51 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10533510 91 - 23771897 AUUGUUGUUGCU---ACUGUCGCUACAGUGCCAUCGUGGCUCUGGCCACU-------GGUCACUUGGCCACUGGACUACAUAGCAUUGGAUACUACCAACC ........((((---(.(((.(...(((.(((.....))).))).(((.(-------((((....))))).))).).))))))))((((......)))).. ( -28.10) >DroSec_CAF1 413113 97 - 1 AUUGUUGUUGCUACUACUGUCGCUAC--UGCCAUCGUGGCUCUGGCCACUG--CAGUGGUCACUUGGCCACUGGACUACAUAGCACUGGAUACUCGCAACC ...((((((((((.....(((((((.--.(((.....)))..)))).....--((((((((....)))))))))))....)))))..........))))). ( -33.40) >DroSim_CAF1 421499 92 - 1 AUUGUUGUUGCUACUACUGUCGCUAC--UGCCAUCGUGGCUCUGGCCACU-------GGUCACUUGGCCACUGGACUCCAUAGCACUGGAUACUAACAACC ...(((((((........(((((((.--.(((.....)))..))))((.(-------((((....))))).)))))((((......))))...))))))). ( -28.60) >DroYak_CAF1 422255 93 - 1 AUUGUUGUUGCC---CCUGUCGCUACAUCGCCAUCGUGGUACUGGCCACUGGCCACUGGUCACUCGGCCACUGGACAACAUAGCACUAGAUACUCC----- ..(((((((...---..............((((..(((((....)))))))))((.(((((....))))).)))))))))................----- ( -26.10) >consensus AUUGUUGUUGCU___ACUGUCGCUAC__UGCCAUCGUGGCUCUGGCCACU_______GGUCACUUGGCCACUGGACUACAUAGCACUGGAUACUACCAACC ..((((..(((((.....(((((......))....((((((.((((((........))))))...))))))..)))....)))))...))))......... (-21.51 = -22.82 + 1.31)

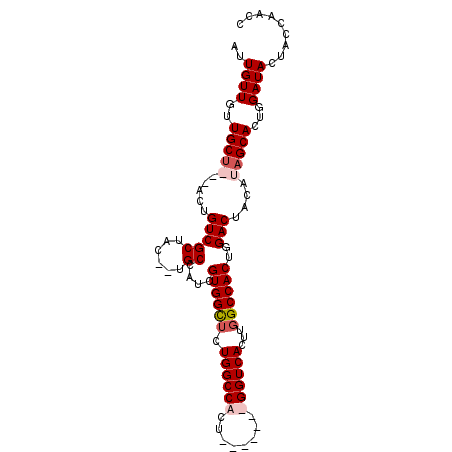
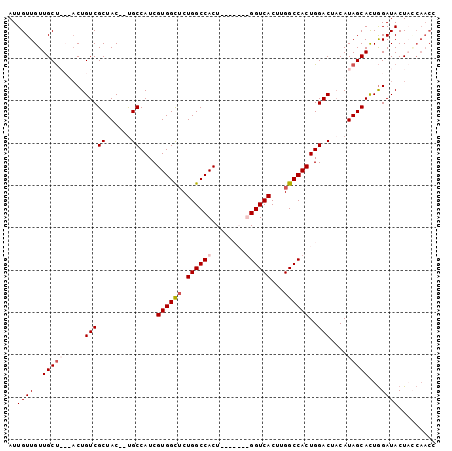
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:25 2006