| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,524,949 – 10,525,094 |
| Length | 145 |
| Max. P | 0.542568 |

| Location | 10,524,949 – 10,525,059 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.12 |
| Energy contribution | -20.79 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
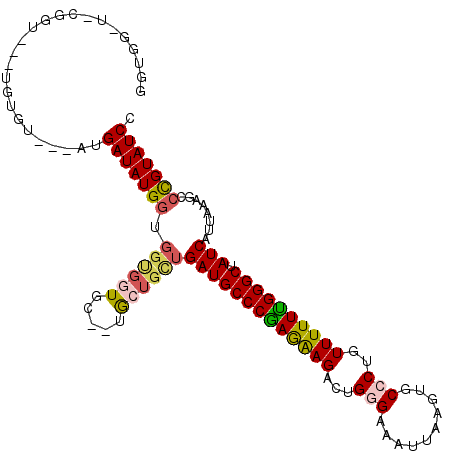
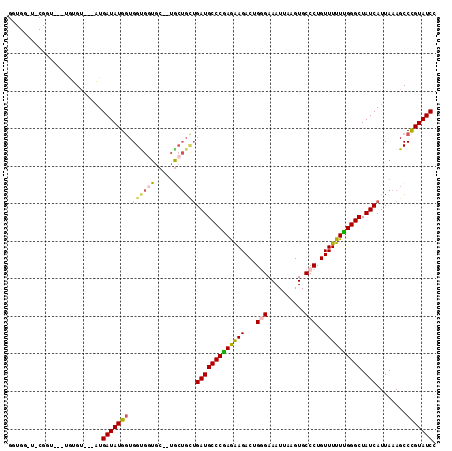
>3L_DroMel_CAF1 10524949 110 - 23771897 GCUGC---UGAUGCC-CAAGAAGACUGGGAAAUUAAGUGCCCUGUUUUUUGGGCUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCCAAAGCAUAUUGAUG ((.((---(((((((-(((((((...(((..........)))..)))))))))).)))).....((((((.....))))))))))..(((.(((((.((....))))))).))) ( -36.40) >DroVir_CAF1 450821 103 - 1 -----AGCAGCUACCCAAAUGAAGCU--GAAAUUAAGUGCCCUCUUU--UAGGCUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGUCUCAUAAAUAUCGCC--AGCAUACUGAUG -----..(((((.(......).))))--).......((((((.(...--((((((........))))))......).))))))................(--((....)))... ( -25.80) >DroGri_CAF1 414337 102 - 1 -----AGCAGCUCCU-CAAUGAAGCU--GAAAUUAAGUGCCCUAUUU--UAGGGUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCC--AGCAUAUUGAUG -----..(((((.(.-....).))))--).......((((((((...--))))))))((((((.((((((.....))))))(((............))).--......)))))) ( -27.60) >DroEre_CAF1 413464 108 - 1 -----UGCGGAUGCC-CGAAGAGACUGGGAAAUUAAGUGCCCUGUUUUUCGGGCUAUCAUUAAAGCCCGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCCAAAGCAUAUUGAUG -----.(((((((((-(((((((...(((..........)))..)))))))))).)))......((((((.....)))))).)))..(((.(((((.((....))))))).))) ( -40.20) >DroWil_CAF1 654512 94 - 1 -------------CU-CCC---AACUG-GAAAUUAAGUGCCCUGUUU--UGGGCUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCCAAAGUAUAUUGAUG -------------.(-((.---....)-))........((((.....--.))))...((((((.((((((.....))))))(((............))).........)))))) ( -21.80) >DroMoj_CAF1 518039 102 - 1 ------GUAGCCGCCCAAAUGAAGCU--GAAAUUAAGUGCCCUGUUU--UAGGCUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCC--AGCAUAUUGAUG ------.................(((--(.......((((((.((..--((((((........))))))......))))))))((............)))--)))......... ( -24.40) >consensus ______GCAGCUGCC_CAAUGAAACU__GAAAUUAAGUGCCCUGUUU__UAGGCUAUCAUUAAAGCCUGUAUCCUGCGGGCGCGCCUCAUAAAUAUCGCC__AGCAUAUUGAUG ....................................((((((.......((((((........))))))........))))))....(((.(((((.((....))))))).))) (-21.12 = -20.79 + -0.33)
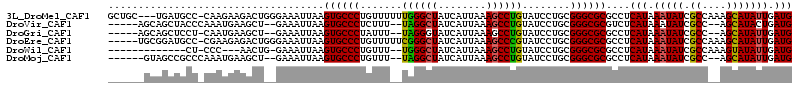
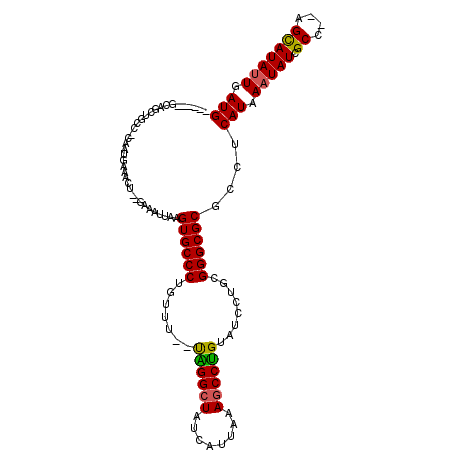
| Location | 10,524,989 – 10,525,094 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10524989 105 - 23771897 GGUGGUU-CGGUGUAUGUGU---AUGAUAUGAUGGCGGUGC--UGC---UGAUGCCCAAGAAGACUGGGAAAUUAAGUGCCCUGUUUUUUGGGCUAUCAUUAAAGCCUGUAUCC .......-............---.......((((.(((.((--(..---((((((((((((((...(((..........)))..)))))))))).))))....)))))))))). ( -29.70) >DroSec_CAF1 404521 94 - 1 ---------------UGUGU---AUGAUAUGGUGGUGGUGC--UGCUGCUGAUGCCCGAGAAGACUGCGAAAUUAAGUGCCCUGUUUUUUGGGCUAUCAUUAAAGCCCGUAUCC ---------------.....---..(((((((.((..(...--..)..))(((((((((((((...(((........)))....)))))))))).)))........))))))). ( -28.30) >DroSim_CAF1 412910 94 - 1 ---------------UAUGU---AUGAUAUGGUGGCGGUGC--UGCUGUUGAUGCCCGAGAAGACUGCGAAAUUAAGUGCCCUGUUUUUUGGGCUAUCAUUAAAGCCCGUAUCC ---------------.....---.......((..((((.((--(.....((((((((((((((...(((........)))....)))))))))).))))....)))))))..)) ( -29.30) >DroEre_CAF1 413504 102 - 1 GGUGGUU-CGGUA--UGUGGGC-AUGAUAUGGUGGUGC--------UGCGGAUGCCCGAAGAGACUGGGAAAUUAAGUGCCCUGUUUUUCGGGCUAUCAUUAAAGCCCGUAUCC ....(((-((...--..)))))-..(((((((....((--------(...(((((((((((((...(((..........)))..)))))))))).))).....)))))))))). ( -34.30) >DroYak_CAF1 413329 112 - 1 GGUGGCUCCGGUA--UGUGUAUGGUGAUAUGGUGGUGCUCCAGUACUGCUGAUGCCCGAAGAGACUGGGAAAUUAAGUGCCCUGUUUUUCGGGCUAUCAUUAAAGCCCGUAUCC ((.....))((((--((.((.((((((((.((..((((....))))..))...((((((((((...(((..........)))..))))))))))))))))))..)).)))))). ( -40.80) >consensus GGUGG_U_CGGU___UGUGU___AUGAUAUGGUGGUGGUGC__UGCUGCUGAUGCCCGAGAAGACUGGGAAAUUAAGUGCCCUGUUUUUUGGGCUAUCAUUAAAGCCCGUAUCC .........................(((((((.((((((.....))))))(((((((((((((...(((..........)))..)))))))))).)))........))))))). (-24.75 = -25.20 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:23 2006