
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,488,663 – 10,488,759 |
| Length | 96 |
| Max. P | 0.757430 |

| Location | 10,488,663 – 10,488,759 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -19.05 |
| Energy contribution | -18.99 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
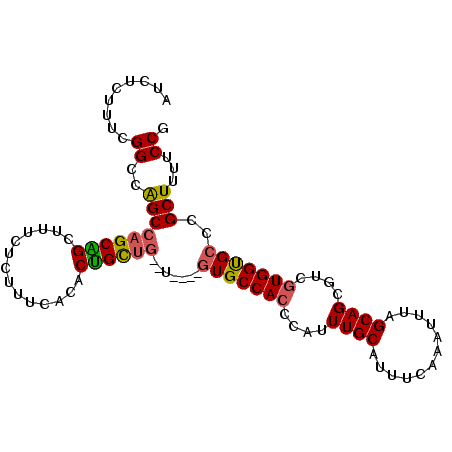
>3L_DroMel_CAF1 10488663 96 + 23771897 AUCUCUUUCGGUCGGCCAGCGGCUUUCUCUUUCUGACCGCCG--U----GUGCCAGCCAUUUGCAUUUCAAAUUUAGCAGCGUCGUGGUGCCCGCUUUUCCG ........(((.((((..(((((..((.......))..))))--)----..))).(((((((((............))))....)))))).)))........ ( -24.00) >DroSec_CAF1 365344 96 + 1 AUCUCUUUCGGCCGGCCAGCAGCUUACUCUUUCUGGCUGCUG--U----GUGCCACCCAUUUGCAUUUCAAAUUCAGCAGCGUCGUGGUGGCCGCUUUCCCA ........(((((.((((((((((..........))))))))--.----))(((((....((((............))))....))))))))))........ ( -32.60) >DroEre_CAF1 375172 98 + 1 AUCUCUUUCGGCCAGCCAGCGGCGUUCUCUUUCACACUGUU----GUGUGUGCCACCCAUUUGCAUUUCAAAUUUAGCAGCGUCGUGGUGCCCGCUUUUCCG .........(((..((((.((((((.......(((((....----)))))(((.....(((((.....)))))...)))))))))))))))).......... ( -25.00) >DroYak_CAF1 376132 102 + 1 AUCUCUUUCGGCCAGCCAGCAGCUUUCUCUUUCACACUGCUGCGUGUGUGUGCCACCCAUUUGCAUUUCAAAUUUAGCAGCCUCGUGGCGCCCGCUUUUCCG ........(((...((((((((..............)))))).))(((.(((((((....((((............))))....))))))).)))....))) ( -27.64) >consensus AUCUCUUUCGGCCAGCCAGCAGCUUUCUCUUUCACACUGCUG__U____GUGCCACCCAUUUGCAUUUCAAAUUUAGCAGCGUCGUGGUGCCCGCUUUUCCG .........((..(((((((((..............)))))).......(((((((....((((............))))....)))))))..)))...)). (-19.05 = -18.99 + -0.06)

| Location | 10,488,663 – 10,488,759 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
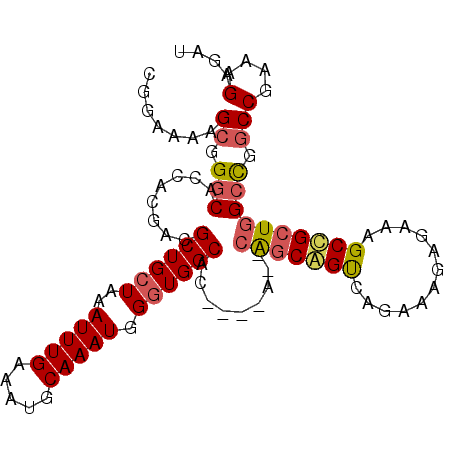
>3L_DroMel_CAF1 10488663 96 - 23771897 CGGAAAAGCGGGCACCACGACGCUGCUAAAUUUGAAAUGCAAAUGGCUGGCAC----A--CGGCGGUCAGAAAGAGAAAGCCGCUGGCCGACCGAAAGAGAU (((...((((..........))))(((..(((((.....))))))))((((.(----(--((((..((.......))..)))).)))))).)))........ ( -27.10) >DroSec_CAF1 365344 96 - 1 UGGGAAAGCGGCCACCACGACGCUGCUGAAUUUGAAAUGCAAAUGGGUGGCAC----A--CAGCAGCCAGAAAGAGUAAGCUGCUGGCCGGCCGAAAGAGAU ........(((((........((..((..(((((.....))))).))..))..----.--(((((((............)))))))...)))))........ ( -32.70) >DroEre_CAF1 375172 98 - 1 CGGAAAAGCGGGCACCACGACGCUGCUAAAUUUGAAAUGCAAAUGGGUGGCACACAC----AACAGUGUGAAAGAGAACGCCGCUGGCUGGCCGAAAGAGAU (((...(((.(((........((..((..(((((.....))))).))..)).(((((----....)))))............))).)))..)))........ ( -30.90) >DroYak_CAF1 376132 102 - 1 CGGAAAAGCGGGCGCCACGAGGCUGCUAAAUUUGAAAUGCAAAUGGGUGGCACACACACGCAGCAGUGUGAAAGAGAAAGCUGCUGGCUGGCCGAAAGAGAU (((...(((.((((((((..(..(((............)))..)..)))))...(((((......)))))............))).)))..)))........ ( -33.80) >consensus CGGAAAAGCGGGCACCACGACGCUGCUAAAUUUGAAAUGCAAAUGGGUGGCAC____A__CAGCAGUCAGAAAGAGAAAGCCGCUGGCCGGCCGAAAGAGAU .......((.(((........((((((..(((((.....))))).)))))).........(((((((............)))))))))).))(....).... (-21.10 = -21.73 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:11 2006