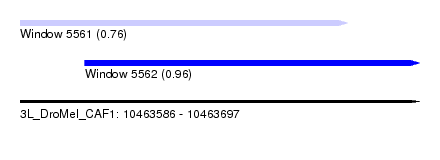
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,463,586 – 10,463,697 |
| Length | 111 |
| Max. P | 0.964753 |

| Location | 10,463,586 – 10,463,677 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.25 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10463586 91 + 23771897 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUAUAUA ...............(((((.((((.......((((((((((....))))))))))............)))------).)))))............. ( -20.91) >DroSec_CAF1 335284 87 + 1 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUA---- ...............(((((.((((.......((((((((((....))))))))))............)))------).))))).........---- ( -20.91) >DroSim_CAF1 352233 87 + 1 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUA---- ...............(((((.((((.......((((((((((....))))))))))............)))------).))))).........---- ( -20.91) >DroEre_CAF1 348884 86 + 1 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACUUAAAUAC------CACCAUACCAUAUAU----- ...............(((((.((((.......((((((((((....))))))))))............)))------).)))))........----- ( -20.91) >DroYak_CAF1 349624 90 + 1 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUACUCGUACCACC--ACCACAUAU----- ..............((((((.(((........((((((((((....))))))))))....(((..........)))..)))--.))))))..----- ( -24.20) >DroAna_CAF1 340728 84 + 1 UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGAAAAUGUUGUCAACACUAAUACUGAAAGAG------CCCCCC--CACAGAC----- ...............(((((.((.........(((((((((......)))))))))......((.....))------...)))--))))...----- ( -16.60) >consensus UGGCAAAACUUUAAAUGUGGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC______CACCAUACCAUAUAU_____ ..((...((.......)).))((((.((....((((((((((....)))))))))).)).))))................................. (-16.31 = -16.25 + -0.06)


| Location | 10,463,604 – 10,463,697 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10463604 93 + 23771897 GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUAUAUAUUUGUAUGUAGACACAUACG ((.((((.......((((((((((....))))))))))............)))------).))...(((((((((......)))))))))......... ( -25.71) >DroSec_CAF1 335302 73 + 1 GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUA--------------------UACG ((.((((.......((((((((((....))))))))))............)))------).))............--------------------.... ( -17.81) >DroSim_CAF1 352251 73 + 1 GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC------CACCAUACUAUAUAUA--------------------UACG ((.((((.......((((((((((....))))))))))............)))------).))............--------------------.... ( -17.81) >DroEre_CAF1 348902 81 + 1 GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACUUAAAUAC------CACCAUACCAUAUAU------------GUGGACACAUACG ((.((((.......((((((((((....))))))))))............)))------).))...((((....------------))))......... ( -21.11) >DroYak_CAF1 349642 85 + 1 GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUACUCGUACCACC--ACCACAUAU------------GUAGACACAUACG ((.((((.(((...((((((((((....))))))))))...((......))..))))))).))--......(((------------((....))))).. ( -20.70) >DroAna_CAF1 340746 74 + 1 GGCGGUAAUGAAAUUGUUGGCAGAAAAUGUUGUCAACACUAAUACUGAAAGAG------CCCCCC--CACAGAC----------------ACACAUAC- ((.(((........(((((((((......)))))))))((.........)).)------))))..--.......----------------........- ( -12.20) >consensus GGCGGUAAUGAAAUUGUUGGCAGCAAAUGUUGUCAACACUAAUACCUAAAUAC______CACCAUACCAUAUAU________________ACACAUACG ...((((.((....((((((((((....)))))))))).)).))))..................................................... (-14.10 = -14.05 + -0.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:09 2006