
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
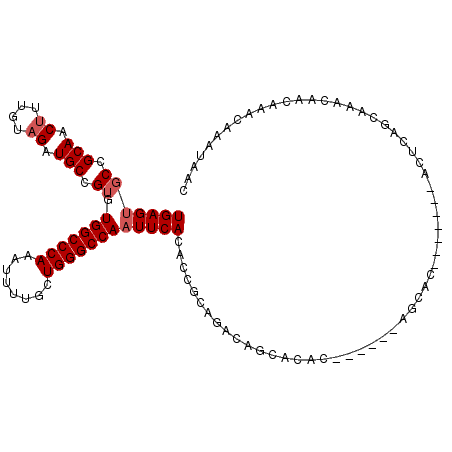
| Location | 10,423,699 – 10,423,806 |
| Length | 107 |
| Max. P | 0.606946 |

| Location | 10,423,699 – 10,423,806 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10423699 107 + 23771897 UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAGACAGCAGAC------AGCACAC--AGCACUCAGCAAACAACAAACAAAUAAC (((((((........(((...(((.((((((((((........)))))).....)))).))).)))((....------.))....--.))))))).................... ( -32.50) >DroSec_CAF1 294665 102 + 1 UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAGACAGCACAC------AGCAC-------ACUCAGCAAACAACAAACAAAUAAC ((((((..((....((((...(((.((((((((((........)))))).....)))).))).)))).....------.)).)-------))))).................... ( -29.10) >DroSim_CAF1 310379 102 + 1 UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAGACAGCACAC------AGCAC-------ACUCAGCAAACAACAAACAAAUAAC ((((((..((....((((...(((.((((((((((........)))))).....)))).))).)))).....------.)).)-------))))).................... ( -29.10) >DroEre_CAF1 306069 107 + 1 UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAGACAGCACAC------AGCAAAC--ACGAUGCAACAAACAGUAAACAAAUAAC ((.((((........(((...(((.((((((((((........)))))).....)))).))).))))))).)------)(((...--....)))..................... ( -26.10) >DroYak_CAF1 308202 115 + 1 UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAAAUAGCAGACACAAAUAGCAAACACACGAAGCAACAAACAACAAACAAAUAAC ....(((.(((.((....)).)))(((((((((((........))))))..........((.....)).................)))))..))).................... ( -24.70) >DroAna_CAF1 300097 87 + 1 UGAGCGCUGCAACUUUGUUGAUGCCG--UGGCCCACAUUUUGCUGGGCCAAUUCACACCCGAGAC--------------------------CACAGCAAACAACAAACAAAUAAG .....((((...(((.(.((.((...--(((((((........)))))))...)))).).)))..--------------------------..)))).................. ( -21.20) >consensus UGAGUGCCGCAACUUUGUAGAUGCCGUGUGGCCCAAAUUUUGCUGGGCCAAUUCACACCGCAGACAGCACAC______AGCAC_______ACUCAGCAAACAACAAACAAAUAAC (((((((.(((.((....)).))).)).(((((((........))))))))))))............................................................ (-17.47 = -17.97 + 0.50)


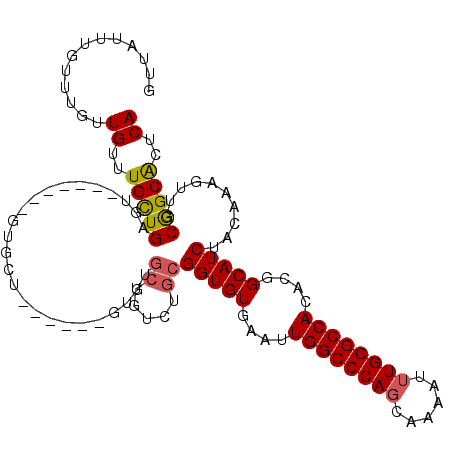
| Location | 10,423,699 – 10,423,806 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -23.98 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10423699 107 - 23771897 GUUAUUUGUUUGUUGUUUGCUGAGUGCU--GUGUGCU------GUCUGCUGUCUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA .....................(((((((--(((((((------((((((.....)))).......((((((((......)))))))).)))))))..........))))))))). ( -39.00) >DroSec_CAF1 294665 102 - 1 GUUAUUUGUUUGUUGUUUGCUGAGU-------GUGCU------GUGUGCUGUCUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA ......((..((((((..(((..((-------(((((------((((((((....)))).......(((((((......))))))))))))))))..))..))).))))))..)) ( -37.60) >DroSim_CAF1 310379 102 - 1 GUUAUUUGUUUGUUGUUUGCUGAGU-------GUGCU------GUGUGCUGUCUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA ......((..((((((..(((..((-------(((((------((((((((....)))).......(((((((......))))))))))))))))..))..))).))))))..)) ( -37.60) >DroEre_CAF1 306069 107 - 1 GUUAUUUGUUUACUGUUUGUUGCAUCGU--GUUUGCU------GUGUGCUGUCUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA ..............(..(((((((...(--((.((((------((((((((....)))).......(((((((......)))))))))))))))...)))....)))))))..). ( -39.10) >DroYak_CAF1 308202 115 - 1 GUUAUUUGUUUGUUGUUUGUUGCUUCGUGUGUUUGCUAUUUGUGUCUGCUAUUUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA .............((..((((((....((((..((((........((((.....)))).(((...((((((((......)))))))))))))))..)))).....))))))..)) ( -35.70) >DroAna_CAF1 300097 87 - 1 CUUAUUUGUUUGUUGUUUGCUGUG--------------------------GUCUCGGGUGUGAAUUGGCCCAGCAAAAUGUGGGCCA--CGGCAUCAACAAAGUUGCAGCGCUCA ........(((((((..(((((((--------------------------((((.((((........)))).((.....))))))))--))))).)))))))............. ( -30.50) >consensus GUUAUUUGUUUGUUGUUUGCUGAGU_______GUGCU______GUGUGCUGUCUGCGGUGUGAAUUGGCCCAGCAAAAUUUGGGCCACACGGCAUCUACAAAGUUGCGGCACUCA .............((..(((((.........................((.....))(((((....((((((((......))))))))....)))))..........)))))..)) (-23.98 = -23.98 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:01 2006