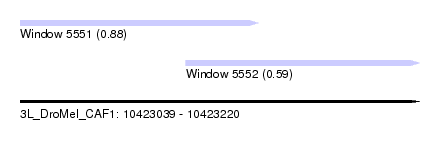
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,423,039 – 10,423,220 |
| Length | 181 |
| Max. P | 0.882783 |

| Location | 10,423,039 – 10,423,147 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -24.21 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10423039 108 + 23771897 UGUGCCACUUUGACCCAGUUGGCU---CCCUAAGCGGGCUAAUGGGGAAAGGUGUAGC-C-AAACAAGCUGCCUGGUAGCCCCAGCUAUUUUUGCUGCCUUUUU-UCAU------UUUUU ..(((((((((..((((.((((((---(.......)))))))))))..)))))(((((-.-......)))))..))))....((((.......)))).......-....------..... ( -32.90) >DroPse_CAF1 319071 107 + 1 UGUGCCACUUUGACCCAGUUGGCUGCCCCCAAAGCGGGCUAAUAGGGAAAGGUGUAGAGC-AAACAAGC-GACUGGCAGUGCCACCGAU-GCGGCUGCUGUUGC-U---------ACUUA ..(((((((((..(((.(((((((((.......)).))))))).))).))))))....))-)....(((-(((.((((((((.......-)).)))))))))))-)---------..... ( -37.60) >DroSim_CAF1 309716 107 + 1 UGUGCCACUUUGACCCAGUUGGCU---CCCUAAGCGGGCUAAUGGGGAAAGGUGUAGC-C-AAACAAGCUGCCUGGCAGCCCCAGCCAUUUUUGCUGCCUUUUU-UCAU------UUUU- ..(((((((((..((((.((((((---(.......)))))))))))..)))))(((((-.-......)))))..))))....((((.......)))).......-....------....- ( -33.70) >DroEre_CAF1 305275 115 + 1 UGUGCCACUUUGACCCAGUUGGCU---UCCUAAGCGGGCUAAUGGGGAAAGGUGUAGC-C-AAACAAGCUGGCUGCUAGCCACAGCCAUUUUUGCUGCCUUUUUCAUUUUUUUGUUUUUU .((..((((((..((((.((((((---(.......)))))))))))..))))))..))-.-(((((((.((((.....))))((((.......)))).............)))))))... ( -32.90) >DroYak_CAF1 307398 115 + 1 UGUGCCACUUUGACCCAGUUGGCU---CCCUAAGCGGGCUAAUGGGGAAAGGUGUAGC-C-AAACAAGCUGGCUGGUAGCCACAGCCAUUUUGGCUGCCUUUUUUUCAUUUUUUUUUUUU ...((((((.......)).)))).---(((.....)))..((((..((((((.(((((-(-(((.....((((((.......)))))).)))))))))))))))..)))).......... ( -40.60) >DroAna_CAF1 299638 94 + 1 UGUGGCACUUUGACCCAGUUGGCU---CCCUAAGCGGGUUAAUGGGGAAAGGUGUAGC-CGAAACAAGCUGGUAUGCAGCCCCUGCGGCU---GCUGCUAU------------------- .((((((((((..((((.((((((---(.......)))))))))))..)))))(((((-((......((((.....)))).....)))))---)).)))))------------------- ( -38.70) >consensus UGUGCCACUUUGACCCAGUUGGCU___CCCUAAGCGGGCUAAUGGGGAAAGGUGUAGC_C_AAACAAGCUGGCUGGCAGCCCCAGCCAUUUUUGCUGCCUUUUU_UCAU______UUUUU .((..((((((..(((((((.(((........))).))))...)))..))))))..))................((((((.............))))))..................... (-24.21 = -24.24 + 0.03)



| Location | 10,423,114 – 10,423,220 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -16.87 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
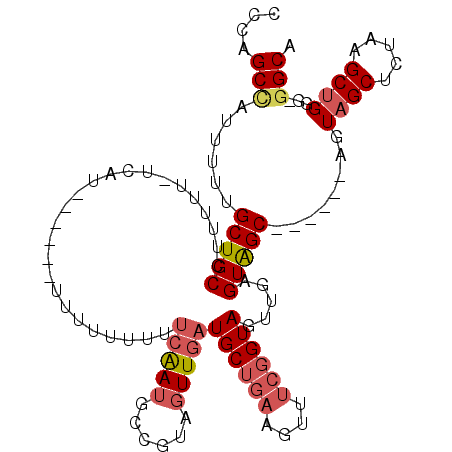
>3L_DroMel_CAF1 10423114 106 + 23771897 CCCAGCUAUUUUUGCUGCCUUUUU-UCAU------UUUUUCUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGCAGUAGCAGUAGCUUUAAGCUGGGGGGCA (((((((.....((((((..(((.-.(..------........(((((......)))))((((((....)))))))..)))..))))))((....))....)))))))..... ( -31.10) >DroSec_CAF1 294084 99 + 1 CCCAGCCAUUUUUGCUGCCUUUUU-UCAU------UUUUUUUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGC------AGUAGCUCUAUGCUGGG-GGCA ((((((.....(((((((..(((.-.(..------........(((((......)))))((((((....)))))))..)))..))------)))))......))))))-.... ( -28.70) >DroSim_CAF1 309791 98 + 1 CCCAGCCAUUUUUGCUGCCUUUUU-UCAU------UUUU-UUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGC------AGUAGCUCUAAGCUGGA-GGCA .(((((.....(((((((..(((.-.(..------....-...(((((......)))))((((((....)))))))..)))..))------)))))......))))).-.... ( -25.50) >DroEre_CAF1 305350 106 + 1 CACAGCCAUUUUUGCUGCCUUUUUCAUUUUUUUGUUUUUUUUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGC------AGUAGCUCUAAGCUGGG-GGCA ....(((....(((((((.........................(((((......)))))((((((....)))))).....)))))------))((((.....))))..-))). ( -24.40) >DroYak_CAF1 307473 112 + 1 CACAGCCAUUUUGGCUGCCUUUUUUUCAUUUUUUUUUUUUUUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGCAGUAGCAGUAGCUCUAAGCUGGG-GGCA ...((((.....))))(((((.....................((((((((((((((....))))......))))).)))))((((((.(((....)))))..))))))-))). ( -26.40) >DroAna_CAF1 299714 89 + 1 CCCUGCGGCU---GCUGCUAU-------------------UUUCAGAUGCUGUACUUGAUGCCAAAGUUGCAGUAGUCGAGUGGCAGUGGCAGUAGCACUA-GCCGCG-AGCU ...(((((((---(.((((((-------------------(.(((..((((..(((((((((.......)))....)))))))))).))).))))))).))-))))))-.... ( -35.20) >consensus CCCAGCCAUUUUUGCUGCCUUUUU_UCAU______UUUUUUUUUCAAUGCCGUAGUUGAUGCUGAAGUUUCGGUAGUUGAGUAGC______AGUAGCUCUAAGCUGGG_GGCA ....(((......(((((.........................(((((......)))))((((((....)))))).....)))))........((((.....))))...))). (-16.87 = -17.82 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:00 2006