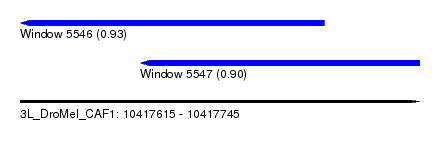
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,417,615 – 10,417,745 |
| Length | 130 |
| Max. P | 0.930444 |

| Location | 10,417,615 – 10,417,714 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.81 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.26 |
| Energy contribution | -23.46 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10417615 99 - 23771897 GCGGCAACAUCAGCAUCUUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU (((((((.....((((.....(((.((((((((((((....)))).)))))))).)))..)))).....))))))).(((((......)))))...... ( -28.90) >DroPse_CAF1 314381 80 - 1 GCG-------------------GCAGCUGGUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU (.(-------------------((.((((.(((((((....)))).))).)))).......((((.(....).))))..............))).)... ( -18.30) >DroSec_CAF1 288645 99 - 1 GCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU (((((((.....((((.....(((.((((((((((((....)))).)))))))).)))..)))).....))))))).(((((......)))))...... ( -28.90) >DroSim_CAF1 304388 99 - 1 GCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU (((((((.....((((.....(((.((((((((((((....)))).)))))))).)))..)))).....))))))).(((((......)))))...... ( -28.90) >DroYak_CAF1 301644 99 - 1 GCAGCAAUAUCAGCAUCUUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU ...((((.....((((.....(((.((((((((((((....)))).)))))))).)))..)))).....)))).((.(((((......))))).))... ( -25.00) >consensus GCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAAAUGCGUCUUUUGUCGUAGUUAAAGUUUAUUGGCCACAUU (((((((.....((((.....(((.((((((((((((....)))).)))))))).)))..)))).....))))))).(((((......)))))...... (-21.26 = -23.46 + 2.20)


| Location | 10,417,654 – 10,417,745 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -20.79 |
| Energy contribution | -22.48 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10417654 91 - 23771897 AGCAACAGGCCAACUAGAGAUAUCUGCGGCAGCGGCAACAUCAGCAUCUUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAA .((.(((((((((.((((....((((.(((((((....)....((((....)))).)))))))))))))))))))))...)).))...... ( -28.30) >DroSec_CAF1 288684 91 - 1 AGCAACAGACCAACUAGCGAUAUCUGCGGCAGCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAA .((.(((((((((.(((....(((((.(((((((....)....((((....)))).)))))))))))))))))))))...)).))...... ( -28.70) >DroSim_CAF1 304427 91 - 1 AGCAACAGGCCAACUAGCGAUAUCUGCGGCAGCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAA .((.(((((((((.(((....(((((.(((((((....)....((((....)))).)))))))))))))))))))))...)).))...... ( -28.10) >DroYak_CAF1 301683 80 - 1 AGCAACAGGCCAACAAGCGA-----------GCAGCAAUAUCAGCAUCUUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAA .((.(((((((((.((((((-----------(((((.......((((....)))).))))))).).))).)))))))...)).))...... ( -21.80) >consensus AGCAACAGGCCAACUAGCGAUAUCUGCGGCAGCGGCAACAUCAGCAUCCUAGUGCAGCUGCUUAGAUUUAUUGGUCUUAAGUAGCAGCAAA .((.(((((((((........(((((.(((((((....)....((((....)))).)))))))))))...)))))))...)).))...... (-20.79 = -22.48 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:55 2006