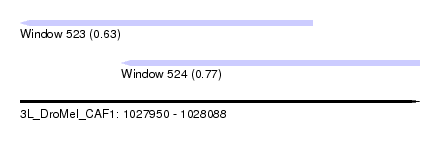
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,027,950 – 1,028,088 |
| Length | 138 |
| Max. P | 0.768947 |

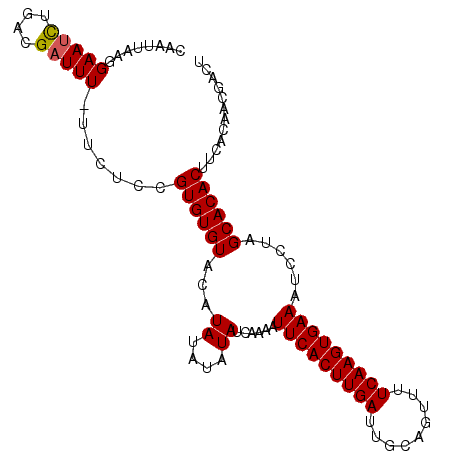
| Location | 1,027,950 – 1,028,051 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -17.25 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1027950 101 - 23771897 AUAUAUAUAACAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUCCACAACGAC----UCGUUGGCUGCGCCUAAGGGCUGAGCAUUGCAGGAU ...............(((((((((.........)))))))))(((((.(((.......(((((..----.)))))(((..(((....)))..)))..)))))))) ( -30.30) >DroVir_CAF1 15942 89 - 1 -----CAUAUUGAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCGUAGCACACUUCAGAGCG-C----UUGUUGGCU-UGCCUAAGGGUUAAGGC-----GGAU -----..........(((((((((.........))))))))).((((......(((.((.(((-(----(....))).-.)))).)))......))-----)).. ( -20.10) >DroGri_CAF1 12524 94 - 1 ACAUACAUAUCGAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCGUAGCACACUUCACAACG-C----UUGUUGGCU-UGCCUAAGGGUUAAACG-----GGUG ..........(((..(((((((((.........))))))))).)))...............((-(----(((((....-.(((....)))..))))-----)))) ( -21.60) >DroEre_CAF1 13325 100 - 1 -UAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUCCACAACGAC----UCGUUGGCUGCGCCUAAGGGCUGAGCAUUGCAGGAU -..............(((((((((.........)))))))))(((((.(((.......(((((..----.)))))(((..(((....)))..)))..)))))))) ( -30.30) >DroMoj_CAF1 12853 94 - 1 AUAUGCAUAUUGAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCGUAGCACACUUCAGAGCG-C----UUGUUGGCC-CGCCUAAGGGUUAAGGC-----GGCU ....((...(((((.(((((((((.........)))))))))...((.....)))))))..((-(----(..((((((-(......))))))))))-----))). ( -26.80) >DroAna_CAF1 9016 104 - 1 -UAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACGACUCACUCGUUGGCAGCGCCUAAGGGCUGAGCAUUGCAGGAU -..............(((((((((.........)))))))))(((((.(((...((((((((((.....)))))).....(((....)))))))...)))))))) ( -30.90) >consensus _UAUACAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACG_C____UCGUUGGCU_CGCCUAAGGGCUAAGCA_____GGAU ...............(((((((((.........)))))))))....((((.........((((.......))))(((...))).....))))............. (-17.25 = -16.28 + -0.97)

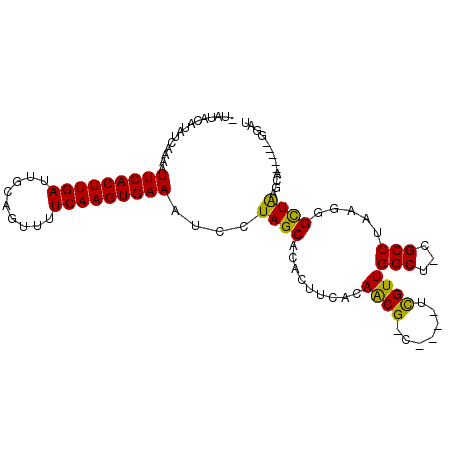
| Location | 1,027,985 – 1,028,088 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1027985 103 - 23771897 CAACUAGGGAAUUUCACGAUUUUUUCUCCGUGUGUACAUAUAUAUAACAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUCCACAACGACU ...((((((.....((((..........))))....................(((((((((.........))))))))).))))))................. ( -20.00) >DroSec_CAF1 10631 102 - 1 CAAUUAGGGAAUCUGACGAUUU-UUCUCCGUGUGUACAUAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACGACU ......(((((((....)))))-....))((((((..(((....))).....(((((((((.........)))))))))......))))))............ ( -18.50) >DroSim_CAF1 10878 102 - 1 GAAUUAAGGAAUCUGACGAUUU-UUCUCCGUGUGUACAUAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACGACU (((...(((((((....)))))-))....((((((..(((....))).....(((((((((.........)))))))))......)))))))))......... ( -19.00) >DroEre_CAF1 13360 100 - 1 CAAUUAAAGAACCUGAUGAUUU-UUCUCCGUGUGUG--UAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUCCACAACGACU ......................-......(((((((--((....))).....(((((((((.........)))))))))......))))))............ ( -16.10) >DroYak_CAF1 11191 100 - 1 CAAUUAAAGAAUCUGAUGAUUU-UUCUCCGUGUGUG--UAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACGACU ......(((((((....)))))-))....(((((((--((....))).....(((((((((.........)))))))))......))))))............ ( -19.60) >consensus CAAUUAAGGAAUCUGACGAUUU_UUCUCCGUGUGUACAUAUAUAUAUCAAAAUUCACUUGAUUGCAGUUUUCAAGUGAAAUCCUAGCACACUUCACAACGACU ........(((((....))))).......((((((...((....))......(((((((((.........)))))))))......))))))............ (-15.62 = -15.66 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:55 2006