
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,371,020 – 10,371,123 |
| Length | 103 |
| Max. P | 0.995015 |

| Location | 10,371,020 – 10,371,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
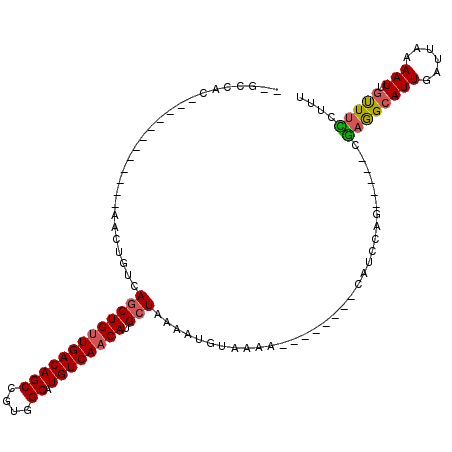
>3L_DroMel_CAF1 10371020 103 + 23771897 --GCCAC------------AACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUACUGGCCAUCCAGCGUCCCGAGGCAUUGAUUAAAAUGGCUUCUUUU --((((.------------....((((((((((((((((....)).)))))))).)))..((((((...(..((((....))))..).....))))))))).....))))....... ( -33.30) >DroVir_CAF1 285265 102 + 1 --ACCUCUAGCUGUUGCCUGUCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUAAAA--------CAUUCAA-----CGAGGCAUUGAUUAAAAUUGUUUUCCUA --.((((((((((((((.((((...(.((((....)))).).)))).).))))).)))).((((.....--------))))...-----.))))....................... ( -25.50) >DroGri_CAF1 263937 102 + 1 --GUUGCAAGCUGUUGCCGUCCUGUCAACUGCUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUAAAA--------CAUUCAA-----CGAGGCAUUGAUUAAAAUUGUUUCCUUU --((((.((....((((((..((((((.....)))))).))(((((((....)))))))....))))..--------..)))))-----)((((((...........)))))).... ( -25.00) >DroWil_CAF1 385188 92 + 1 UUGCUUU------------AGCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUUAAA--------CAUCUUG-----CAAGGCAUUGAUUAAAAUUGUUUUUAUU .((((((------------.((....(((((((((((((....)).)))))))).)))...(((.....--------)))...)-----)))))))..................... ( -24.40) >DroMoj_CAF1 322283 102 + 1 --GCCACUAGCUGUUGCCUGUCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUUAAAUGUAAAA--------CAUUCAG-----CGAAGCAUUGAUUAAAAUUGCUUUCCUU --.......((((.............(((((((((((((....)).)))))))).)))..((((.....--------)))))))-----)((((((...........)))))).... ( -26.80) >DroAna_CAF1 250974 100 + 1 --GCUGA------------AACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCCGGCCAUCCAC---CAAGAGGCAUUGAUUAAAAUUUUUCCUUUU --(((((------------.....)))))((((((((((....)).)))))))).((......))...((((((..........---.....))))))................... ( -26.86) >consensus __GCCAC____________AACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUAAAA________CAUCCAG_____CGAGGCAUUGAUUAAAAUUGUUUCCUUU ..........................(((((((((((((....)).)))))))).)))................................((((((((......))).))))).... (-17.76 = -17.68 + -0.08)

| Location | 10,371,020 – 10,371,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10371020 103 - 23771897 AAAAGAAGCCAUUUUAAUCAAUGCCUCGGGACGCUGGAUGGCCAGUAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUU------------GUGGC-- .......(((((.......(((((...((((..((((....))))..)))).)))))(((((.((((((((.((....))))))))))))))).....------------)))))-- ( -35.70) >DroVir_CAF1 285265 102 - 1 UAGGAAAACAAUUUUAAUCAAUGCCUCG-----UUGAAUG--------UUUUACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGACAGGCAACAGCUAGAGGU-- ......................((((((-----(((..((--------(((......(((((.((((((((.((....))))))))))))))).)))))..)))).....)))))-- ( -30.90) >DroGri_CAF1 263937 102 - 1 AAAGGAAACAAUUUUAAUCAAUGCCUCG-----UUGAAUG--------UUUUACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAGCAGUUGACAGGACGGCAACAGCUUGCAAC-- ...(....)..................(-----(((((((--------.....)).)))))).((((((((.((....))))))))))((((.((((..(....)..))))))))-- ( -27.90) >DroWil_CAF1 385188 92 - 1 AAUAAAAACAAUUUUAAUCAAUGCCUUG-----CAAGAUG--------UUUAACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGCU------------AAAGCAA .....................(((.(((-----(..((((--------.....))))(((((.((((((((.((....)))))))))))))))..)).------------)).))). ( -24.90) >DroMoj_CAF1 322283 102 - 1 AAGGAAAGCAAUUUUAAUCAAUGCUUCG-----CUGAAUG--------UUUUACAUUUAAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGACAGGCAACAGCUAGUGGC-- ...(((.(((...........))))))(-----(((..((--------(((........(((.((((((((.((....))))))))))))).......))))).)))).......-- ( -27.16) >DroAna_CAF1 250974 100 - 1 AAAAGGAAAAAUUUUAAUCAAUGCCUCUUG---GUGGAUGGCCGGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUU------------UCAGC-- ......................(((....)---)).((((((((((((.(((((((......))))))).)))))..)))))))....(((((.....------------)))))-- ( -30.10) >consensus AAAAAAAACAAUUUUAAUCAAUGCCUCG_____CUGAAUG________UUUAACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGAU____________GAAGC__ .........................................................(((((.((((((((.((....)))))))))))))))........................ (-18.63 = -18.52 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:37 2006