
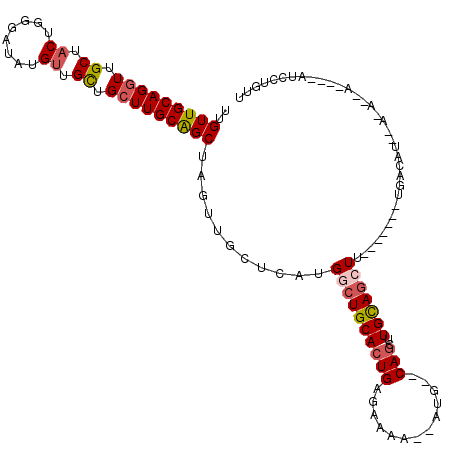
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,370,521 – 10,370,630 |
| Length | 109 |
| Max. P | 0.999044 |

| Location | 10,370,521 – 10,370,630 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10370521 109 + 23771897 UUGUUGCAGGUUGCUACUGGGAUAUGAUGUUGCUUGCAGCUAGUUGCUUAUGAGUGCACUGAGAAAA--AUG--CAGCUUGCAGCUC-------UGACAUACAAAUGAUGUUAUCCUGUU ..((((((((((((..............((((....))))((((.((........))))))......--..)--)))))))))))..-------((((((.(....)))))))....... ( -29.50) >DroSec_CAF1 244254 97 + 1 UAGUUGCAGGUUGCUACUGGGAUAUGUUGCUGCUUGCAGCUAGUUGCUCAUGGCUGCACUGAGAGAA--AUG--CAGCUUGCAGCUU-------UUACAU------------AUCCUGUU .(((.((.....)).)))(((((((((.((((....)))).((((((....(((((((.(......)--.))--))))).)))))).-------..))))------------)))))... ( -37.40) >DroSim_CAF1 254932 97 + 1 UUGUUGCAGGUUGCUACUGGGAUAUGUUGCUGCUUGCAGCUAGUUGCUCAUGGCUGCACUGAGAGAA--AUG--CAGCUUGCAGCUU-------UGACAU------------AUCCUGUU ..((.((.....)).)).((((((((((((((....)))).((((((....(((((((.(......)--.))--))))).)))))).-------.)))))------------)))))... ( -38.00) >DroEre_CAF1 259107 109 + 1 UUGUUGCAGGUUGCUACUGGGAUAUGUUGCUGCUUGCAGCUAGUUGGCCGUGACUGCACUGCCAAAA--ACGCACAGUAUGUAGAUUGGGUUAC-AACAUAAAAAAUA--------UGUU ..(((((((((.((.((........)).)).)))))))))...(((((.(((....))).))))).(--((.(.((((......))))))))..-((((((.....))--------)))) ( -30.70) >DroYak_CAF1 256747 120 + 1 UUGUUGCAGGUUGCUACUGGGAUAUGUUGCUGGUUGCGGCUAGUUGCUCAUGGCUGCACUGCCAAAAAAAAUCACACCUUGCAGCUAAGGUUACUUACAUUAACAAUACUUUAUCCUGUU ..(((((((((.((.((........)).))((((((((((((........))))))))..))))...........))).))))))((((....))))....................... ( -29.80) >consensus UUGUUGCAGGUUGCUACUGGGAUAUGUUGCUGCUUGCAGCUAGUUGCUCAUGGCUGCACUGAGAAAA__AUG__CAGCUUGCAGCUU_______UGACAU__A_A__A____AUCCUGUU ..(((((((((.((.((........)).)).)))))))))...........((((((((((.............)))..))))))).................................. (-20.96 = -21.68 + 0.72)

| Location | 10,370,521 – 10,370,630 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -12.55 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10370521 109 - 23771897 AACAGGAUAACAUCAUUUGUAUGUCA-------GAGCUGCAAGCUG--CAU--UUUUCUCAGUGCACUCAUAAGCAACUAGCUGCAAGCAACAUCAUAUCCCAGUAGCAACCUGCAACAA ..((((.........((((.....))-------))(((((.((.((--(((--(......)))))))).....(((......)))..................)))))..))))...... ( -21.30) >DroSec_CAF1 244254 97 - 1 AACAGGAU------------AUGUAA-------AAGCUGCAAGCUG--CAU--UUCUCUCAGUGCAGCCAUGAGCAACUAGCUGCAAGCAGCAACAUAUCCCAGUAGCAACCUGCAACUA ....((((------------((((..-------..(((((..((((--(((--(......))))))))..(((((.....))).)).))))).)))))))).(((.((.....)).))). ( -35.60) >DroSim_CAF1 254932 97 - 1 AACAGGAU------------AUGUCA-------AAGCUGCAAGCUG--CAU--UUCUCUCAGUGCAGCCAUGAGCAACUAGCUGCAAGCAGCAACAUAUCCCAGUAGCAACCUGCAACAA ....((((------------((((..-------..(((((..((((--(((--(......))))))))..(((((.....))).)).))))).))))))))..((((....))))..... ( -34.90) >DroEre_CAF1 259107 109 - 1 AACA--------UAUUUUUUAUGUU-GUAACCCAAUCUACAUACUGUGCGU--UUUUGGCAGUGCAGUCACGGCCAACUAGCUGCAAGCAGCAACAUAUCCCAGUAGCAACCUGCAACAA ....--------.........((((-(((.......((((...........--..(((((.(((....))).)))))...((((....))))...........)))).....))))))). ( -24.90) >DroYak_CAF1 256747 120 - 1 AACAGGAUAAAGUAUUGUUAAUGUAAGUAACCUUAGCUGCAAGGUGUGAUUUUUUUUGGCAGUGCAGCCAUGAGCAACUAGCCGCAACCAGCAACAUAUCCCAGUAGCAACCUGCAACAA ..((((.................((((....))))(((((..((((((.........((((((((........)).))).)))((.....))..))))))...)))))..))))...... ( -28.50) >consensus AACAGGAU____U__U_U__AUGUAA_______AAGCUGCAAGCUG__CAU__UUUUCUCAGUGCAGCCAUGAGCAACUAGCUGCAAGCAGCAACAUAUCCCAGUAGCAACCUGCAACAA ..((((.............................(((((..((((.............)))))))))............(((((..................)))))..))))...... (-12.55 = -13.67 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:35 2006