
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,369,478 – 10,369,615 |
| Length | 137 |
| Max. P | 0.996992 |

| Location | 10,369,478 – 10,369,583 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -31.27 |
| Energy contribution | -32.03 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10369478 105 + 23771897 CAUGCGUCUUUGAUUGCUUUGGCUUUGGCUUUGGCGUUGGCUUUUCAGUGGCCAUCGCUGC-----------UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((.((((...(((..(((...(((((.(((((.......))))).))))).-----------.)))..)))..))))(((((.....)))))..)))).))))).. ( -39.30) >DroSec_CAF1 243213 105 + 1 CAUGCGUCUUUGAUUGCUUUGGCUUUGGCUUUGCCGUUGUCUUUUCAGUGGCCAUCGCUGC-----------UCUAUUGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((.((((...(((..(((....((.((.(((........))).)).))...-----------.)))..)))..))))(((((.....)))))..)))).))))).. ( -32.20) >DroSim_CAF1 253899 105 + 1 CAUGCGUCUUUGAUUGCUUUGGCUUUGGCUUUGCCGUUGGCUUUUCAGUGGCCAUCGCUGC-----------UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((.((((...(((..(((....((...(((((.......)))))..))...-----------.)))..)))..))))(((((.....)))))..)))).))))).. ( -35.20) >DroEre_CAF1 257994 105 + 1 CAUGCGUCUUUGAUUGCUUUGGCUUUGGCUUUCGCGUUGGCUUUUCAGUGGCCAUCGCUGC-----------UCCAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((.((((...(((..(((.....(((.(((((.......))))).)))...-----------.)))..)))..))))(((((.....)))))..)))).))))).. ( -39.90) >DroYak_CAF1 255616 116 + 1 CAUGCGUCUUUGAUUGCUUUAGCUUUGGCUUUGGCGUUGGCUUUUCAGUGGCCAUCGCUGCACCAACACUGCACCAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((...((....((...(((..(((((.(((((.......))))).)))((((.......)))).))..)))..))..(((((.....))))))))))).))))).. ( -38.80) >consensus CAUGCGUCUUUGAUUGCUUUGGCUUUGGCUUUGGCGUUGGCUUUUCAGUGGCCAUCGCUGC___________UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAAC ..(((((.((((.((((...(((..(((....((((.(((((.......))))).))))..............)))..)))..))))(((((.....)))))..)))).))))).. (-31.27 = -32.03 + 0.76)


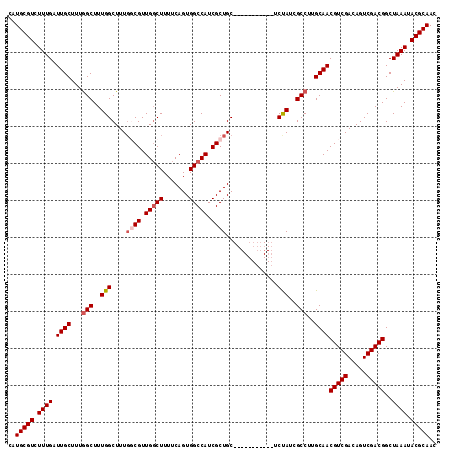
| Location | 10,369,515 – 10,369,615 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10369515 100 + 23771897 UGGCUUUUCAGUGGCCAUCGCUGC-----------UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGAGGGCCACGCCCUCAAAAAAGC ..((((((...(((((...((.((-----------......))...))...((((.....)))))))))..................(((((((...))))))).)))))) ( -30.90) >DroSec_CAF1 243250 100 + 1 UGUCUUUUCAGUGGCCAUCGCUGC-----------UCUAUUGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGAUGGCCACGCCCUCAAAAAAGC ..........(((((((((((((.-----------....((((.......(((((.....))))).........))))......))))))))))))).............. ( -34.79) >DroSim_CAF1 253936 100 + 1 UGGCUUUUCAGUGGCCAUCGCUGC-----------UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGAGGGCCACGCCCUCAAAAAAGC ..((((((...(((((...((.((-----------......))...))...((((.....)))))))))..................(((((((...))))))).)))))) ( -30.90) >DroEre_CAF1 258031 99 + 1 UGGCUUUUCAGUGGCCAUCGCUGC-----------UCCAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGCGGGCCACGCCCUCAAA-AAGC ..((((((..((((((...(((((-----------...........))).(((((.....)))))))......((((.(......).)))))))))).......))-)))) ( -30.30) >DroYak_CAF1 255653 111 + 1 UGGCUUUUCAGUGGCCAUCGCUGCACCAACACUGCACCAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGAGGGCCACGCCCUCAAAAAAGC .(((......((((((...(.((((.......)))).).(((((......(((((.....)))))((.......)).........))))).)))))))))........... ( -32.70) >consensus UGGCUUUUCAGUGGCCAUCGCUGC___________UCUAUCGCCUUGCAACGUCGACAGUCGACGGCUAAAUACGCAACACCAUCGGUGAGGGCCACGCCCUCAAAAAAGC ..........((((((.((((((...........................(((((.....)))))((.......))........)))))).)))))).............. (-28.42 = -28.62 + 0.20)

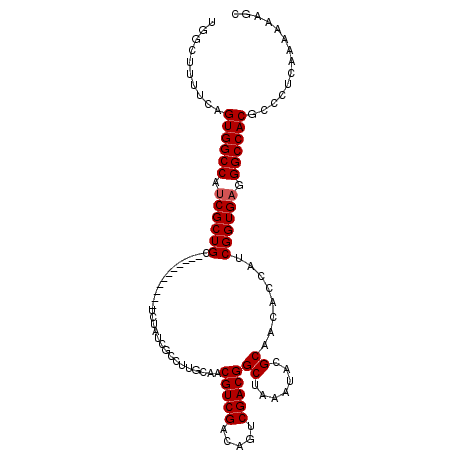
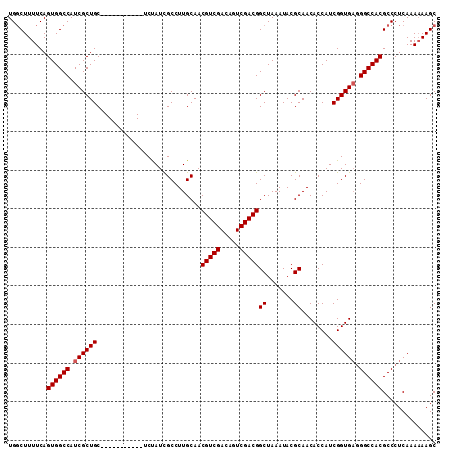
| Location | 10,369,515 – 10,369,615 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10369515 100 - 23771897 GCUUUUUUGAGGGCGUGGCCCUCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCGAUAGA-----------GCAGCGAUGGCCACUGAAAAGCCA (((((((((((.(((..((((........)).))..))).))))(((((((.(((((...(((((...))))).))-----------.))))))))))....))))))).. ( -37.10) >DroSec_CAF1 243250 100 - 1 GCUUUUUUGAGGGCGUGGCCAUCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCAAUAGA-----------GCAGCGAUGGCCACUGAAAAGACA (((((....)))))((((((((((((...)))(((((..........((((.....))))(((((...)))))...-----------)))))))))))))).......... ( -38.00) >DroSim_CAF1 253936 100 - 1 GCUUUUUUGAGGGCGUGGCCCUCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCGAUAGA-----------GCAGCGAUGGCCACUGAAAAGCCA (((((((((((.(((..((((........)).))..))).))))(((((((.(((((...(((((...))))).))-----------.))))))))))....))))))).. ( -37.10) >DroEre_CAF1 258031 99 - 1 GCUU-UUUGAGGGCGUGGCCCGCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCGAUGGA-----------GCAGCGAUGGCCACUGAAAAGCCA ((((-(((.((.(((..(((((......))).))..))).....(((((((.(((.(..((((((...))))))..-----------)))))))))))..))))))))).. ( -41.20) >DroYak_CAF1 255653 111 - 1 GCUUUUUUGAGGGCGUGGCCCUCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCGAUGGUGCAGUGUUGGUGCAGCGAUGGCCACUGAAAAGCCA (((((((((((.(((..((((........)).))..))).))))(((((((.((((((((((((((.........))))))))))..)))))))))))....))))))).. ( -43.10) >consensus GCUUUUUUGAGGGCGUGGCCCUCACCGAUGGUGUUGCGUAUUUAGCCGUCGACUGUCGACGUUGCAAGGCGAUAGA___________GCAGCGAUGGCCACUGAAAAGCCA ((((......))))((((((.(((((...)))(((((..........((((.....))))(((((...)))))..............))))))).)))))).......... (-32.32 = -32.36 + 0.04)

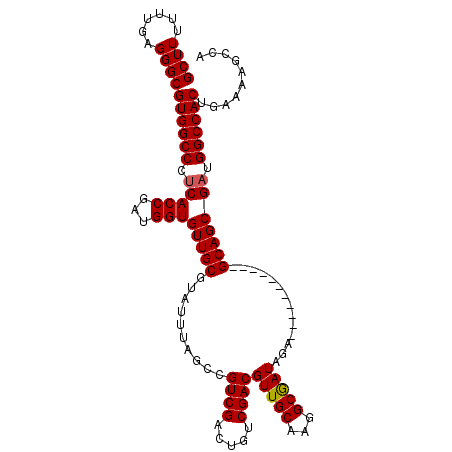
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:33 2006