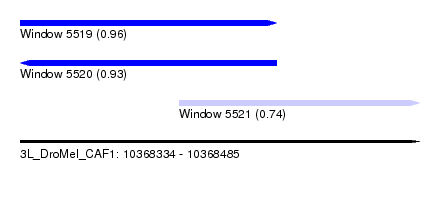
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,368,334 – 10,368,485 |
| Length | 151 |
| Max. P | 0.956447 |

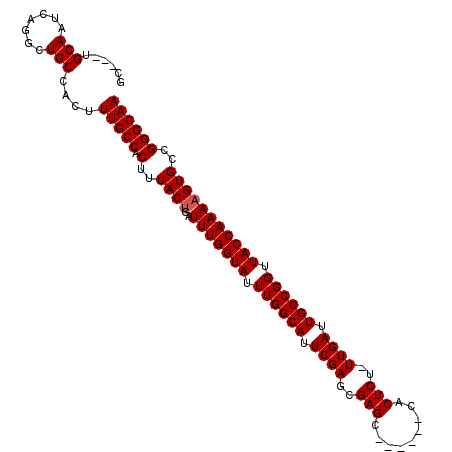
| Location | 10,368,334 – 10,368,431 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -28.77 |
| Energy contribution | -28.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
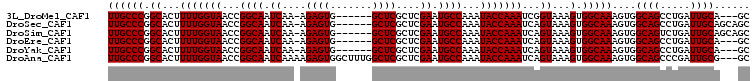
>3L_DroMel_CAF1 10368334 97 + 23771897 UUGCCCGGCACUUUUGGUAACCGGCAAUCAA-AGAGUG------GCUCGCUCGAAUGCCAAAUACCAAAUCGGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA---GC .....(((.((.....))..)))(((((((.-.....(------(((.(((....(((((..((((.....))))...)))))....)))))))))))))).---.. ( -31.30) >DroSec_CAF1 242082 100 + 1 UUGCCCGGCACUUUUGGUAACCGGCAAUCAA-AGAGUG------GCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCAGCAGC ((((((.((...(((((((...((((.((..-.(((((------...))))))).))))...)))))))...))...).))))).((.((((.....)))).))... ( -29.70) >DroSim_CAF1 252768 100 + 1 UUGCCCGGCACUUUUGGUAACCGGCAAUCAA-AGAGUG------GCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGUCUGAUUGCAGCAGC ((((((.((...(((((((...((((.((..-.(((((------...))))))).))))...)))))))...))...).))))).((.(((((...))))).))... ( -30.40) >DroEre_CAF1 256762 97 + 1 UUGCCCGGCACUUUUGGUAACCGGCAAUCAA-AGAGUG------GCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA---GC ((((((.((...(((((((...((((.((..-.(((((------...))))))).))))...)))))))...))...).)))))....((((.....)))).---.. ( -28.10) >DroYak_CAF1 254548 97 + 1 UUGCCCGGCACUUUUGGUAACCGGCAAUCAA-AGAGUG------GCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA---GC ((((((.((...(((((((...((((.((..-.(((((------...))))))).))))...)))))))...))...).)))))....((((.....)))).---.. ( -28.10) >DroAna_CAF1 248505 104 + 1 UUGCCCGGCACUUUUGGUAACCGGCAAUCAAAAGAGUGGCUUUGGCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGCCCGAUUGCG---GC ((((((.((...(((((((...((((.((....(((((((....)).))))))).))))...)))))))...))...).))))).((.((((.....)))).---)) ( -34.50) >consensus UUGCCCGGCACUUUUGGUAACCGGCAAUCAA_AGAGUG______GCUCGCUCGAAUGCCAAAUACCAAAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA___GC ((((((.((...(((((((...((((.((....((((.......))))....)).))))...)))))))...))...).)))))....((((.....))))...... (-28.77 = -28.77 + 0.00)
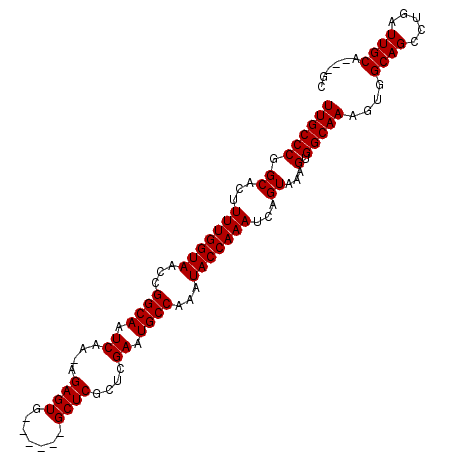
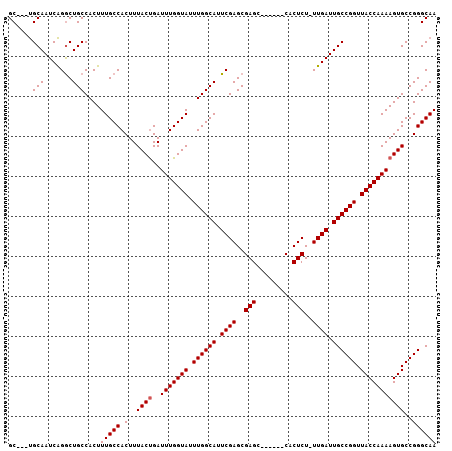
| Location | 10,368,334 – 10,368,431 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -28.73 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10368334 97 - 23771897 GC---UGCAAUCAGGCUGCCACUUUGCCACUUUACCGAUUUGGUAUUUGGCAUUCGAGCGAGC------CACUCU-UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((---((((((((((((((.....(((((...((((.....))))..))))).....)).)))------).....-.)))))))((((.((.....))))))))).. ( -33.90) >DroSec_CAF1 242082 100 - 1 GCUGCUGCAAUCAGGCUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGC------CACUCU-UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((.(((.......))).))....(((((.(..((((..(((((((.((((((.(((((.(((.------..))))-)))).)))))).)))))))))))..)))))) ( -34.10) >DroSim_CAF1 252768 100 - 1 GCUGCUGCAAUCAGACUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGC------CACUCU-UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((..(((....)))...))....(((((.(..((((..(((((((.((((((.(((((.(((.------..))))-)))).)))))).)))))))))))..)))))) ( -32.70) >DroEre_CAF1 256762 97 - 1 GC---UGCAAUCAGGCUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGC------CACUCU-UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((---.((......)).))....(((((.(..((((..(((((((.((((((.(((((.(((.------..))))-)))).)))))).)))))))))))..)))))) ( -33.10) >DroYak_CAF1 254548 97 - 1 GC---UGCAAUCAGGCUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGC------CACUCU-UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((---.((......)).))....(((((.(..((((..(((((((.((((((.(((((.(((.------..))))-)))).)))))).)))))))))))..)))))) ( -33.10) >DroAna_CAF1 248505 104 - 1 GC---CGCAAUCGGGCUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGCCAAAGCCACUCUUUUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ((---((((((((((((((.....(((((...((((.....))))..))))).....)).))))((((.....)))))))))))((((.((.....))))))))).. ( -35.40) >consensus GC___UGCAAUCAGGCUGCCACUUUGCCACUUUACUGAUUUGGUAUUUGGCAUUCGAGCGAGC______CACUCU_UUGAUUGCCGGUUACCAAAAGUGCCGGGCAA ......(((.......)))....(((((.(..((((..(((((((.((((((.((((..(((.........)))..)))).)))))).)))))))))))..)))))) (-28.73 = -28.90 + 0.17)

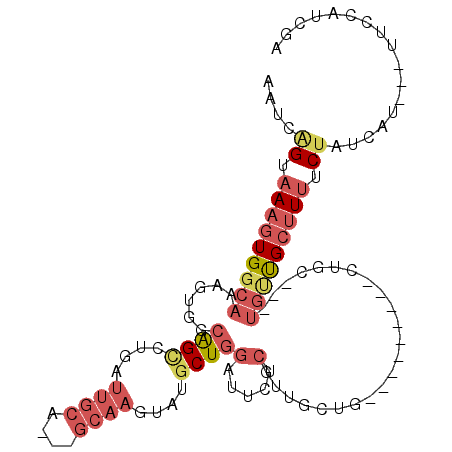
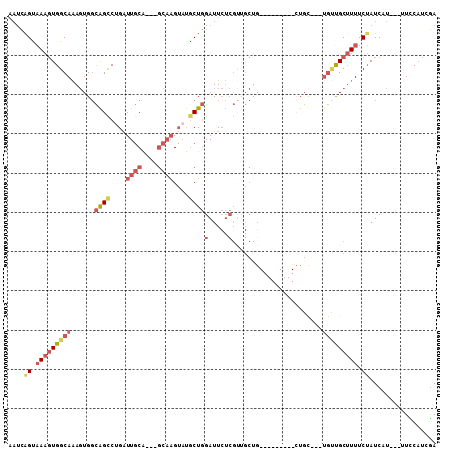
| Location | 10,368,394 – 10,368,485 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.60 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -8.68 |
| Energy contribution | -10.27 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10368394 91 + 23771897 AAUCGGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA---GCAAGUAUGCUGGAUUCUCGUUGCUG---------CUGC---UGUUGCUUUUCUAUCAU---UUCCAUCGA ....((.((((..(((.((..(((((..((.(.((---(((....))))).).))..)))))..---------))..---)))..)))).)).....---......... ( -32.90) >DroGri_CAF1 261641 83 + 1 ---UAGCCAACUGGCAGAUUGCUGGUCUGCUUUCA------UGAUUGCUCGAUU-----G---G---------CUGCUGUUGCCGCAUUUCAAUCCUUGAUUUCCUCAA ---.(((..((..((.....))..))..))).(((------.(((((...((((-----(---(---------(.......)))).))).)))))..)))......... ( -19.60) >DroSec_CAF1 242142 94 + 1 AAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCAGCAGCAAGUAUGCUGGAUUCUCGUUGCUG---------UUGC---UGUUGCUUUUCUAUCAU---UUCCAUCGA ....((.((((..(((.((..(((((..((.(.(((((.......))))).).))..)))))..---------))..---)))..)))).)).....---......... ( -30.60) >DroSim_CAF1 252828 94 + 1 AAUCAGUAAAGUGGCAAAGUGGCAGUCUGAUUGCAGCAGCAAGUAUGCUGGAUUCUCGUUGCUG---------UUGC---UGUUGCUUUUCUAUCAU---UUCCAUCGA ....((.((((..(((..(..(((((..((.(.(((((.......))))).).)).....))))---------)..)---)))..)))).)).....---......... ( -27.10) >DroYak_CAF1 254608 100 + 1 AAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA---GCAAGUAUGCUGGAUUCUCGUGGCUGCUGCUGCUGCUGU---UGUUGCUUUUCUAUCAU---UUCCAUCGA ....(((((((..(((.((..((((((.((.(.((---(((....))))).).))....))))))..))..)))..)---).)))))..........---......... ( -36.10) >DroAna_CAF1 248572 82 + 1 AAUCAGUAAAGUGGCAAAGUGGCAGCCCGAUUGCG---GCAAGUACACUGGAUUCUCGCUGC---------------------CGCUUUUCUAUCAU---UUUCAUCGA ..........((((.(((((((((((..((.(.((---(........))).).))..)))))---------------------)))))).))))...---......... ( -25.10) >consensus AAUCAGUAAAGUGGCAAAGUGGCAGCCUGAUUGCA___GCAAGUAUGCUGGAUUCUCGUUGCUG_________CUGC___UGUUGCUUUUCUAUCAU___UUCCAUCGA ....((.(((((((((......((((....((((....))))....))))(.....).......................))))))))).))................. ( -8.68 = -10.27 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:31 2006