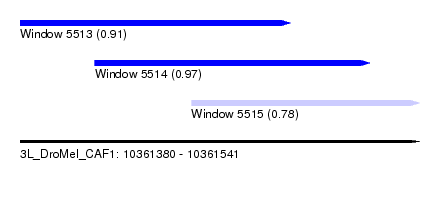
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,361,380 – 10,361,541 |
| Length | 161 |
| Max. P | 0.968634 |

| Location | 10,361,380 – 10,361,489 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10361380 109 + 23771897 --------GCUCUAUACU-AUAUUUGUACGAAGUAAAUA-UCACCAUAUUU-AUUUUCCAAGUGUACUGACCGCUGUCCACCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUA --------..........-(((((((...((((((((((-(....))))))-))))).)))))))((..(((.((((((.......)))))).((....)))))..))............ ( -26.00) >DroSec_CAF1 235047 89 + 1 ------------------------------AAGGAUGUAUUCACCAUAUUU-ACUUUCCGAGUGUACUGACCGCUGUCCGCCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUA ------------------------------..(((.(((...........)-))..)))((((.(((..(((.((((((.......)))))).((....)))))..)))..))))..... ( -26.10) >DroSim_CAF1 245421 89 + 1 ------------------------------AAGGAUGUAUUCACCAUAUUU-ACUUUCCGAGUGUACUGACCGCUGUCCGCCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUA ------------------------------..(((.(((...........)-))..)))((((.(((..(((.((((((.......)))))).((....)))))..)))..))))..... ( -26.10) >DroEre_CAF1 249525 111 + 1 UGAAAUGUGUACUUAACUAUUA------UUGAGUAAAUU-UCAU-ACGAUU-UUUUUCUGAGUGUACUGACCGCUGUCCACCUUCAGGAAAGGAGAAUGCUGGCUGGUAAAACUCCUAUA ((((((...(((((((......------))))))).)))-))).-......-..((((((((((.((........)).)))..)))))))(((((..((((....))))...)))))... ( -26.50) >DroYak_CAF1 247151 112 + 1 -----UAUACACUUUA-UGUUAUUUGCUUUCAGUAAAUG-UCAU-AUAUUUAAUUUUCCAAGUGUACUGACCGCUGUCCACCUUCAGGAAAGGAGAAUGCUGGUUGGUAAAACUCCGAUA -----...((((((((-((.(((((((.....)))))))-.)))-).............))))))((..((((((.(((.((....))...))).)..)).)))..))............ ( -23.71) >consensus ___________CU__A_U__UA________AAGUAAAUA_UCACCAUAUUU_AUUUUCCGAGUGUACUGACCGCUGUCCACCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUA ...........................................................((((.((((((((.((((((.......)))))).((....))))))))))..))))..... (-16.60 = -17.60 + 1.00)

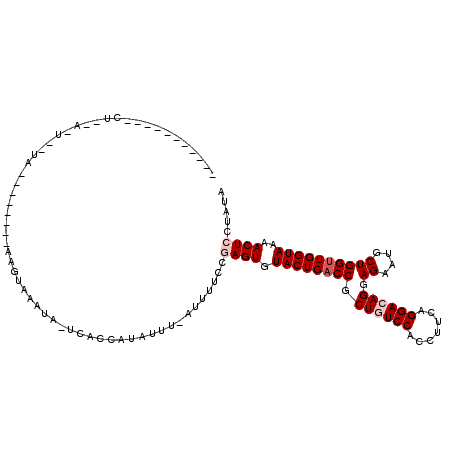
| Location | 10,361,410 – 10,361,521 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -28.42 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10361410 111 + 23771897 UCACCAUAUUU-AUUUUCCAAGUGUACUGACCGCUGUCCACCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGACUUGCCAAUAAUGUCAUUGUGCUGCAA ...........-........((..((.((((..((((((.......))))))..........((((((((...(((......))).))))))))...)))).))..)).... ( -28.40) >DroSec_CAF1 235057 111 + 1 UCACCAUAUUU-ACUUUCCGAGUGUACUGACCGCUGUCCGCCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAA ...........-........((..((.((((..((((((.......))))))..........(((((((..(((((......))))))))))))...)))).))..)).... ( -32.90) >DroSim_CAF1 245431 111 + 1 UCACCAUAUUU-ACUUUCCGAGUGUACUGACCGCUGUCCGCCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAA ...........-........((..((.((((..((((((.......))))))..........(((((((..(((((......))))))))))))...)))).))..)).... ( -32.90) >DroEre_CAF1 249558 110 + 1 UCAU-ACGAUU-UUUUUCUGAGUGUACUGACCGCUGUCCACCUUCAGGAAAGGAGAAUGCUGGCUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAA ....-....((-((((((((((((.((........)).)))..)))))))))))...(((.((((((((..(((((......))))))))))(((((...))))))))))). ( -29.30) >DroYak_CAF1 247184 111 + 1 UCAU-AUAUUUAAUUUUCCAAGUGUACUGACCGCUGUCCACCUUCAGGAAAGGAGAAUGCUGGUUGGUAAAACUCCGAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAA ((((-((((((........))))))).)))..((..(((.......))).((.(.(((((..(((((((..((((((....)))))))))))))...).)))).).)))).. ( -26.90) >consensus UCACCAUAUUU_AUUUUCCGAGUGUACUGACCGCUGUCCACCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAA ....................((..((.((((..((((((.......))))))..........(((((((..(((((......))))))))))))...)))).))..)).... (-28.42 = -29.22 + 0.80)

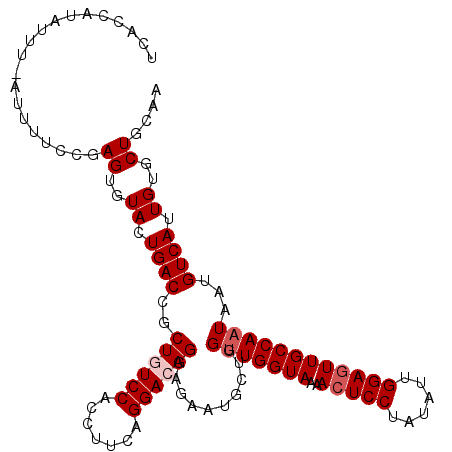
| Location | 10,361,449 – 10,361,541 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10361449 92 + 23771897 CCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGACUUGCCAAUAAUGUCAUUGUGCUGCAACUGCCAUGCCAAAAGACAGC .((((......))))...((((((((((((...(((......))).)))))))).......(((.((.((....))...)))))....)))) ( -22.20) >DroSec_CAF1 235096 92 + 1 CCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAACUGCCAUGCCAAAAGACAGC .((((......))))...(((((((((((..(((((......)))))))))))).......(((.((.((....))...)))))....)))) ( -25.90) >DroSim_CAF1 245470 91 + 1 CCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAACUG-CAUGCCAAAAGACAAC .((((......)))).......(((((((..(((((......))))))))))))..((((.(((.(((((....)-)).)))))..)))).. ( -27.00) >DroEre_CAF1 249596 92 + 1 CCUUCAGGAAAGGAGAAUGCUGGCUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAACUGCCAUGCCAAAAGACAGC .((((......)))).....(((((((((.((((((......))))))((((((......)))).))......))))).))))......... ( -25.70) >DroYak_CAF1 247223 92 + 1 CCUUCAGGAAAGGAGAAUGCUGGUUGGUAAAACUCCGAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAACUGCCAUGCCAAAAGACAGC .((((......))))...(((((((((((..((((((....))))))))))))).......(((.((.((....))...)))))....)))) ( -25.40) >consensus CCUUCAGGACAGGAGAAUGCUGGUUGGUAAAACUCCUAUAUUGGAGUUGCCAAUAAUGUCAUUGUGCUGCAACUGCCAUGCCAAAAGACAGC ....(((..(((.(.(((((..(((((((..(((((......))))))))))))...).)))).).)))...)))................. (-22.02 = -22.82 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:25 2006