
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,317,224 – 10,317,318 |
| Length | 94 |
| Max. P | 0.716857 |

| Location | 10,317,224 – 10,317,318 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.77 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711178 |
| Prediction | RNA |
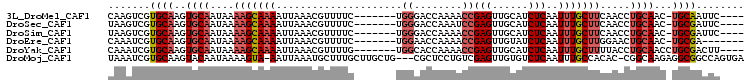
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10317224 94 + 23771897 CAAGUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUC-------UGGGACCAAAACCGAGUUGCAUCUCAAUUUGCUUCAACCUGCAAC-UGCAAUUC---- .......((((..((((....(((((((.............-------.((........))(((......)))..))))))).....))))..-))))....---- ( -20.70) >DroSec_CAF1 191657 94 + 1 UAAGUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUC-------UGGGACCAAAUCCGAGUUGCAUCUCAAUUUGCUUCAACCUGCAAC-UGCGAUUC---- .......((((..((((....(((((((.......((...(-------(.(((.....))).))..)).......))))))).....))))..-))))....---- ( -20.24) >DroSim_CAF1 197149 94 + 1 UAAGUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUC-------UGGGACCAAAACCGAGUUGCAUCUCAAUUUGCUUCAACCUGCAAC-UGCGAUUC---- .......((((..((((....(((((((.............-------.((........))(((......)))..))))))).....))))..-))))....---- ( -20.00) >DroEre_CAF1 203423 91 + 1 CAAAUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUC-------UGGAACCAAAACCGAGUUGUAUCUCAAUUUGCUUGGAACUGCAAC-UGCGA------- .......((((..((((....(((((((.......((((..-------((....)))))).(((......)))..))))))).....))))..-)))).------- ( -20.00) >DroYak_CAF1 201089 95 + 1 CAAAUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUG-------UGGCACCAAAACCGAGUUGCAUCUCAAUUUGCUUUUACCUGCAACCUGCGACUU---- ......(((((..((((.((((((((((.......((((((-------......)))))).(((......)))..))))))))))..))))...))).))..---- ( -25.00) >DroMoj_CAF1 254161 101 + 1 UAAAUCGUGCAAGUACAAUAAAAGUA-AAUUAAAUGCUUUGCUUGCUG---CGCUCCUGUCGAGUUGUGUCUCAAUUUGCCACAC-CGGCAAGAGGCGGCCAGUGA ......(((....)))....((((((-.......))))))(((.((((---(.(((.(((((.((.(((.(.......).)))))-))))).)))))))).))).. ( -29.00) >consensus CAAAUCGUGCAAGUGCAAUAAAAGCAAAAUUAAACGUUUUC_______UGGGACCAAAACCGAGUUGCAUCUCAAUUUGCUUCAACCUGCAAC_UGCGAUUC____ .......((((..((((....(((((((.....................((........))(((......)))..))))))).....))))...))))........ (-13.71 = -14.77 + 1.06)


| Location | 10,317,224 – 10,317,318 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -15.27 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
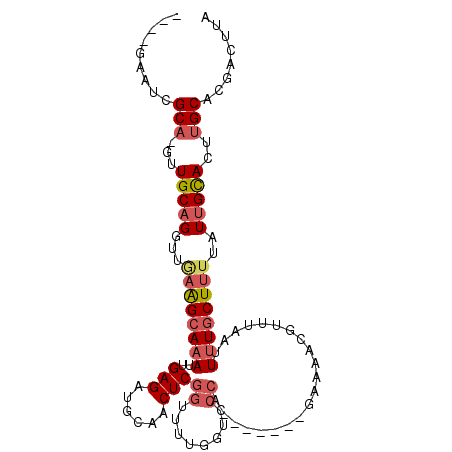
>3L_DroMel_CAF1 10317224 94 - 23771897 ----GAAUUGCA-GUUGCAGGUUGAAGCAAAUUGAGAUGCAACUCGGUUUUGGUCCCA-------GAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGACUUG ----....((((-(.(((((...(((((((((((((......)))))(((((....))-------)))..........))))))))..))))).)))))....... ( -25.50) >DroSec_CAF1 191657 94 - 1 ----GAAUCGCA-GUUGCAGGUUGAAGCAAAUUGAGAUGCAACUCGGAUUUGGUCCCA-------GAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGACUUA ----.....(((-(.(((((...((((((((...(((((...((.((((...)))).)-------)....)))))...))))))))..))))).))))........ ( -26.60) >DroSim_CAF1 197149 94 - 1 ----GAAUCGCA-GUUGCAGGUUGAAGCAAAUUGAGAUGCAACUCGGUUUUGGUCCCA-------GAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGACUUA ----.....(((-(.(((((...(((((((((((((......)))))(((((....))-------)))..........))))))))..))))).))))........ ( -25.20) >DroEre_CAF1 203423 91 - 1 -------UCGCA-GUUGCAGUUCCAAGCAAAUUGAGAUACAACUCGGUUUUGGUUCCA-------GAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGAUUUG -------..(((-(.((((((...((((((((((((......)))))(((((....))-------)))..........)))))))..)))))).))))........ ( -24.20) >DroYak_CAF1 201089 95 - 1 ----AAGUCGCAGGUUGCAGGUAAAAGCAAAUUGAGAUGCAACUCGGUUUUGGUGCCA-------CAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGAUUUG ----(((((((((((.((((.((((((((((.((((......))))((((((......-------)))))).......))))))))))))))))))))..))))). ( -32.00) >DroMoj_CAF1 254161 101 - 1 UCACUGGCCGCCUCUUGCCG-GUGUGGCAAAUUGAGACACAACUCGACAGGAGCG---CAGCAAGCAAAGCAUUUAAUU-UACUUUUAUUGUACUUGCACGAUUUA ......((.((((((((.((-(((((...........))).)).)).)))))).)---).))..((((.(((..(((..-.....))).)))..))))........ ( -22.90) >consensus ____GAAUCGCA_GUUGCAGGUUGAAGCAAAUUGAGAUGCAACUCGGUUUUGGUCCCA_______GAAAACGUUUAAUUUUGCUUUUAUUGCACUUGCACGACUUA .........(((...(((((...((((((((..(((......)))((........)).....................))))))))..)))))..)))........ (-15.27 = -16.22 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:05 2006