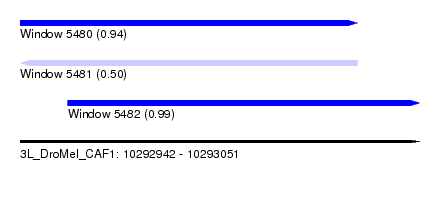
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,292,942 – 10,293,051 |
| Length | 109 |
| Max. P | 0.991889 |

| Location | 10,292,942 – 10,293,034 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10292942 92 + 23771897 CGUUUACAACAUGACUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUUGUU .....(((((((((.((......)).)))...((((((...((((((((((.((....))....))))))))))..))))))...)))))). ( -22.30) >DroVir_CAF1 201079 83 + 1 -----ACAAAGUAA----CUUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUGAAAUGAUAAUUUGUGUGCAGCCAAGUGUUUUUGUAGCU -----((((((..(----((((((....(((((....)))))(((((((((.............))))))))))))))))..)))))).... ( -22.32) >DroPse_CAF1 180810 76 + 1 ---------CA---CAGCAACGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGGUUUUUU---- ---------((---(.......(((......))).......((((((((((.((....))....))))))))))....))).......---- ( -14.50) >DroGri_CAF1 181297 83 + 1 -----CCAAAGUAA----CUUGACAAUCAAAUGCAUUUAUUUUGCAUAUAAUUGAAAUGAUAAUUUGUGUGCAACAAAGUGUUUUUGUAUAU -----.........----.........((((.((((((...((((((((((.............))))))))))..)))))).))))..... ( -15.22) >DroEre_CAF1 178466 92 + 1 CGUUUACAACAUGGCUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUU ...((((((....(((.....)))........((((((...((((((((((.((....))....))))))))))..))))))..)))))).. ( -21.10) >DroAna_CAF1 179155 80 + 1 ------------GGUUGGCUUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUGGUU ------------.........(((...((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..))) ( -19.60) >consensus _____ACAACAUGACUGGCUUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUU ...........................((((.((((((...((((((((((.............))))))))))..)))))).))))..... (-15.19 = -15.22 + 0.03)


| Location | 10,292,942 – 10,293,034 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -14.48 |
| Consensus MFE | -6.95 |
| Energy contribution | -7.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10292942 92 - 23771897 AACAACAAAAGCACUUGACUGCACACAAAUUAUCAUUUGAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCAGCCCAGUCAUGUUGUAAACG .((((((........((((((..(.(((((....)))))(((((.(((((.......))))).)))))....)..))))))))))))..... ( -19.10) >DroVir_CAF1 201079 83 - 1 AGCUACAAAAACACUUGGCUGCACACAAAUUAUCAUUUCAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCAAG----UUACUUUGU----- ....(((((...(((((((((....))............(((((.(((((.......))))).)))))))))))----)...)))))----- ( -17.40) >DroPse_CAF1 180810 76 - 1 ----AAAAAACCACUUGACUGCACACAAAUUAUCAUUUGAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCGUUGCUG---UG--------- ----.......(((..(((.((...(((((....)))))(((((.(((((.......))))).))))))).)))...)---))--------- ( -13.50) >DroGri_CAF1 181297 83 - 1 AUAUACAAAAACACUUUGUUGCACACAAAUUAUCAUUUCAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGUCAAG----UUACUUUGG----- ....(((((.....)))))......((((.((......((((((.(((((.......))))).)))))).....----.)).)))).----- ( -12.10) >DroEre_CAF1 178466 92 - 1 AACUACAAAAGCACUUGACUGCACACAAAUUAUCAUUUGAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCAGCCCAGCCAUGUUGUAAACG ...(((((.....((.(.((((...(((((....)))))(((((.(((((.......))))).)))))).))).).)).....))))).... ( -13.90) >DroAna_CAF1 179155 80 - 1 AACCACAAAAGCACUUGACUGCACACAAAUUAUCAUUUGAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCAAGCCAACC------------ .............((((...((...(((((....)))))(((((.(((((.......))))).)))))))))))......------------ ( -10.90) >consensus AACUACAAAAGCACUUGACUGCACACAAAUUAUCAUUUGAAUUAUAUGCAAAAUAAAUGCAUUUGAUUGCCAAGCCAGUCAUGUUGU_____ ....................((...(((((....)))))(((((.(((((.......))))).)))))))...................... ( -6.95 = -7.15 + 0.20)

| Location | 10,292,955 – 10,293,051 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -26.04 |
| Energy contribution | -28.52 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10292955 96 + 23771897 ACUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUUGUUGCCAGCUUGCAGUUGUG ((((((((((((((((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..))))))))))..)))).... ( -37.60) >DroPse_CAF1 180812 74 + 1 -CAGCAACGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGGUUUUUU--------------------- -..((....)).........(((((...((((((((((.((....))....))))))))))..))))).......--------------------- ( -13.80) >DroSim_CAF1 174422 96 + 1 GCUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUUGCCAGCUUGCAGUUGUG ((.(((((((((((((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..)))))))))))))....... ( -38.10) >DroEre_CAF1 178479 96 + 1 GCUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUUGCCAGUUUGCAGUUGUG ((.(((((((((((((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..)))))))))))))....... ( -35.60) >DroYak_CAF1 176554 96 + 1 GCUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUUGCCAGCUUGCAGUUGUG ((.(((((((((((((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..)))))))))))))....... ( -38.10) >DroAna_CAF1 179156 94 + 1 GUUGGCUUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUGGUUGUC--UUUGUAGUUGUA ....((..((((((((...((((((...((((((((((.((....))....))))))))))..)))))).....))))))))--...))....... ( -26.60) >consensus GCUGGGCUGGCAAUCAAAUGCAUUUAUUUUGCAUAUAAUUCAAAUGAUAAUUUGUGUGCAGUCAAGUGCUUUUGUAGUUGCCAGCUUGCAGUUGUG ...(((((((((((((((.((((((...((((((((((.((....))....))))))))))..)))))).))))..)))))))))))......... (-26.04 = -28.52 + 2.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:54 2006