
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,288,003 – 10,288,095 |
| Length | 92 |
| Max. P | 0.795851 |

| Location | 10,288,003 – 10,288,095 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -10.32 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10288003 92 + 23771897 GCCAUGACAAUGAACCAAACUGUCUUUUCCAGUUUUGAAAGUUCAUAAGUUUUU----UAAACUUUUUUACUUUU-------UUUAACUCCCAAAACUCCCAU .....((((...........))))......((((..((((((....(((((...----..)))))....))))))-------...)))).............. ( -10.50) >DroSec_CAF1 162975 87 + 1 GCCAUGACAAUGAACCAAACUGUCUUUUUCAAUUUUGUAAGUUCAUAAGUAUAUUAAUUAUACUUAUUUACUUUU-------UUUAACUCCCAA--------- .....((((...........))))..............((((..(((((((((.....)))))))))..))))..-------............--------- ( -11.70) >DroSim_CAF1 169431 89 + 1 GCCAUGACAAUGAACCAAACUGUCUUUUCCAAUUUUUUAAGUUCAUAAGUAUUU----UAUACUUAUUUACUUU--------UUUAACUCCCAAAUC--CUUU .....((((...........))))..............((((..((((((((..----.))))))))..)))).--------...............--.... ( -8.70) >DroYak_CAF1 171205 83 + 1 GCCAUGACAAUGAACCAAAUUGUCUUUUCGAAUUUUUCAAGUUCAUAAGUUCAA----GUGACAUAUUUACUCUUAUAGUAGUUUAC---------------- .....((((((.......)))))).....(((((.....)))))(((((....(----((((.....))))))))))..........---------------- ( -10.40) >consensus GCCAUGACAAUGAACCAAACUGUCUUUUCCAAUUUUGUAAGUUCAUAAGUAUAU____UAUACUUAUUUACUUUU_______UUUAACUCCCAA_________ .....((((...........))))..............((((..((((((((.......))))))))..)))).............................. ( -6.99 = -7.42 + 0.44)

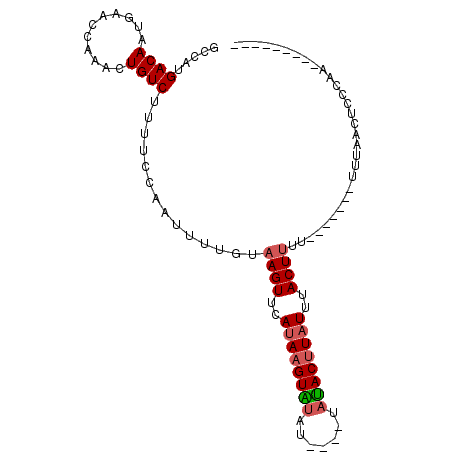

| Location | 10,288,003 – 10,288,095 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -14.52 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10288003 92 - 23771897 AUGGGAGUUUUGGGAGUUAAA-------AAAAGUAAAAAAGUUUA----AAAAACUUAUGAACUUUCAAAACUGGAAAAGACAGUUUGGUUCAUUGUCAUGGC .....(((((((..((((...-------(.((((...........----....)))).).))))..)))))))......((((((.......))))))..... ( -18.36) >DroSec_CAF1 162975 87 - 1 ---------UUGGGAGUUAAA-------AAAAGUAAAUAAGUAUAAUUAAUAUACUUAUGAACUUACAAAAUUGAAAAAGACAGUUUGGUUCAUUGUCAUGGC ---------(((..((((...-------........(((((((((.....))))))))).))))..)))..........((((((.......))))))..... ( -15.30) >DroSim_CAF1 169431 89 - 1 AAAG--GAUUUGGGAGUUAAA--------AAAGUAAAUAAGUAUA----AAAUACUUAUGAACUUAAAAAAUUGGAAAAGACAGUUUGGUUCAUUGUCAUGGC ....--(((...(((......--------.((((..((((((((.----..))))))))..))))...((((((.......))))))..)))...)))..... ( -14.00) >DroYak_CAF1 171205 83 - 1 ----------------GUAAACUACUAUAAGAGUAAAUAUGUCAC----UUGAACUUAUGAACUUGAAAAAUUCGAAAAGACAAUUUGGUUCAUUGUCAUGGC ----------------......((((.....)))).....((((.----.(((....(((((((........((.....))......)))))))..))))))) ( -10.44) >consensus _________UUGGGAGUUAAA_______AAAAGUAAAUAAGUAUA____AAAAACUUAUGAACUUAAAAAAUUGGAAAAGACAGUUUGGUUCAUUGUCAUGGC ..............................((((..((((((((.......))))))))..))))..............((((((.......))))))..... (-10.75 = -11.12 + 0.38)

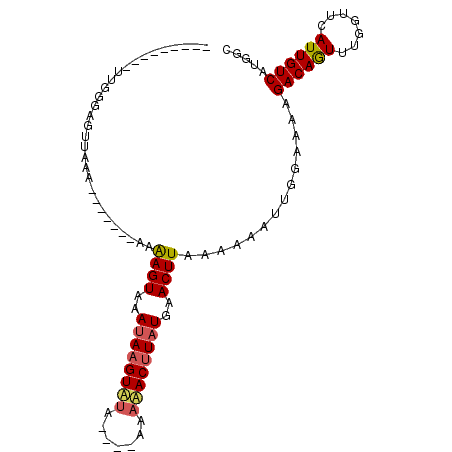
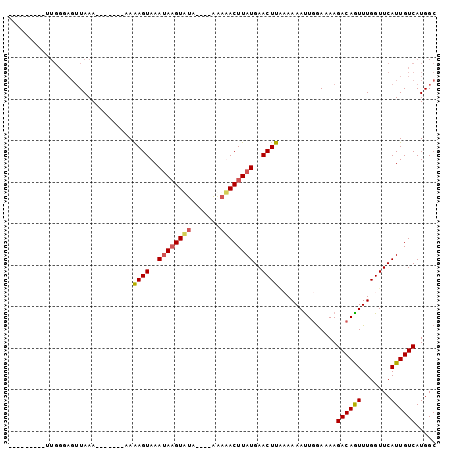
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:48 2006