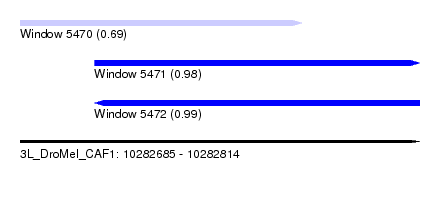
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,282,685 – 10,282,814 |
| Length | 129 |
| Max. P | 0.990473 |

| Location | 10,282,685 – 10,282,776 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -15.52 |
| Consensus MFE | -9.05 |
| Energy contribution | -9.58 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10282685 91 + 23771897 AUACAUCGAUCACAACACACGC---------CGGCUAAUUUGCAUAAAUUUUUGCGAAUUUACUCGCA--UUUU-----GUUUUUUUUUUGCACUCGCAAAUUCUUU ....................((---------.((..((((((((........))))))))..)).)).--....-----........(((((....)))))...... ( -15.50) >DroSec_CAF1 157697 93 + 1 AUACAUCGAUCACAACACACGC---------CGGCUAAUUUGCAUAAAUUUUUGCAAAA-AACCCGCA--UUUCUUU--UUUUUUUUUUUGCACGCGCAAAUUCUUU ....................((---------.((....((((((........)))))).-..)).)).--.......--........(((((....)))))...... ( -13.30) >DroSim_CAF1 163788 89 + 1 AUGCAUCGAUCACAACACACGC---------CGGCUAAUUUGCAUAAAUUUUUGCGAAUUUACUCGCA--UUUCUUU--U-----UUUUUGCACGCGCAAAUUCUUU ....................((---------.((..((((((((........))))))))..)).)).--.......--.-----..(((((....)))))...... ( -15.50) >DroEre_CAF1 168090 97 + 1 AUACAUCGAUCACAACACACGC---------CGGCUAAUUUGCAUAAAUU-UUGCGAAUUUACUCGCUAUUUUCUUUUUUUUUUUUUUUUGCACGCGCAAAUUCUUU ....................((---------.((..((((((((......-.))))))))..)).))....................(((((....)))))...... ( -14.70) >DroYak_CAF1 165864 87 + 1 AUACAUCGAUCAGAACACACGC---------CGGCUAAUUUGCAUAAAUUUUUGCGAGUUUACUCGCU-----------UUUUUUUAUUUGCACGCGCAAAUUCUUU ...........((((.....((---------.((..((((((((........))))))))..)).)).-----------........(((((....))))))))).. ( -16.10) >DroAna_CAF1 169733 94 + 1 AUACAUCGAUCACAACACACACACACACUGGCAGCUAAUUUGCAUAAAUUUUUGCAAUUUCGUUGGAG-------------UUUUUUGUUGGUUGUGCAAAUUUUUU ..........((((((.((.(((...(((..((((.((.(((((........))))).)).)))).))-------------)....))))))))))).......... ( -18.00) >consensus AUACAUCGAUCACAACACACGC_________CGGCUAAUUUGCAUAAAUUUUUGCGAAUUUACUCGCA__UUUC_____UUUUUUUUUUUGCACGCGCAAAUUCUUU ...............................(((..((((((((........))))))))...))).....................(((((....)))))...... ( -9.05 = -9.58 + 0.53)

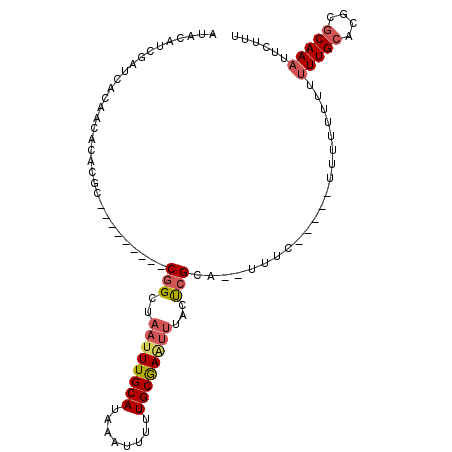
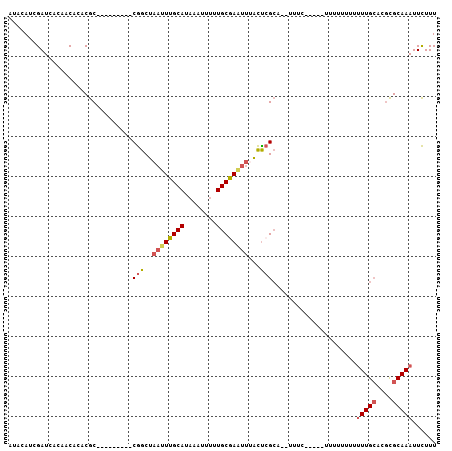
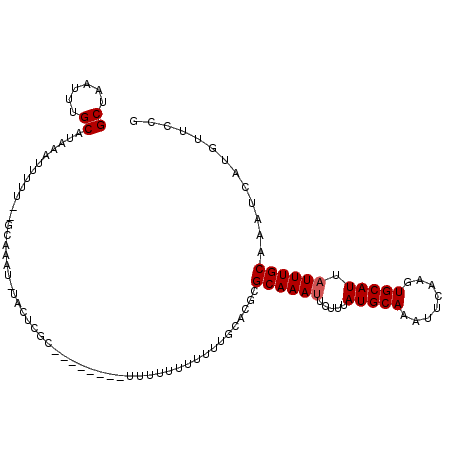
| Location | 10,282,709 – 10,282,814 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -10.85 |
| Energy contribution | -11.02 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10282709 105 + 23771897 GCUAAUUUGCAUAAAUUUUU--GCGAAUUUACUCGCAUUUU---GUUUUUUUUUUGCACUCGCAAAUUCUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCG (((((((((((((((....(--((((......)))))....---........(((((....)))))...))))))))))))..)))(((.....)))............. ( -24.10) >DroVir_CAF1 189926 90 + 1 GCUAAUUUGCAUAAAUUGUUUGGCAGUU-U-----------UU-UUUUUUUUUUUG-----GCAAAC--CUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCC ((((((((((((((...(((((.(((..-.-----------..-.........)))-----.)))))--.)))))))))))..)))(((.....)))............. ( -20.14) >DroPse_CAF1 170981 82 + 1 GCUAAUUUGCAUAUUUUUUU--------------------C---UUUUUUUUUUUG-----GCAAAUUCUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUGCA ((.((((((((((....(((--------------------(---(..........)-----).))).....))))))))))....((((.....)))).........)). ( -12.80) >DroSec_CAF1 157721 107 + 1 GCUAAUUUGCAUAAAUUUUU--GCAAAA-AACCCGCAUUUCUUUUUUUUUUUUUUGCACGCGCAAAUUCUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCG (((((((((((((((....(--((....-.....)))...............(((((....)))))...))))))))))))..)))(((.....)))............. ( -19.30) >DroYak_CAF1 165888 101 + 1 GCUAAUUUGCAUAAAUUUUU--GCGAGUUUACUCGCU-------UUUUUUUAUUUGCACGCGCAAAUUCUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCG (((((((((((((((.....--(((((....))))).-------.......((((((....))))))..))))))))))))..)))(((.....)))............. ( -26.80) >DroAna_CAF1 169766 99 + 1 GCUAAUUUGCAUAAAUUUUU--GCAAUUUCGUUGGAG---------UUUUUUGUUGGUUGUGCAAAUUUUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCG (((((((((((((((..(((--(((...(((......---------........)))...))))))...))))))))))))..)))(((.....)))............. ( -21.94) >consensus GCUAAUUUGCAUAAAUUUUU__GCAAAU_UACUCGC________UUUUUUUUUUUGCACGCGCAAAUUCUUUAUGCAAAUUCAAGUGCAUUAUUUGCAAAUCAUGUUCCG ((......))...................................................((((((.....(((((........))))).))))))............. (-10.85 = -11.02 + 0.17)
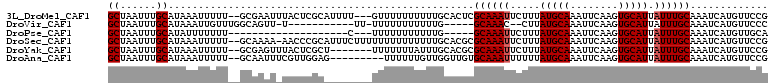

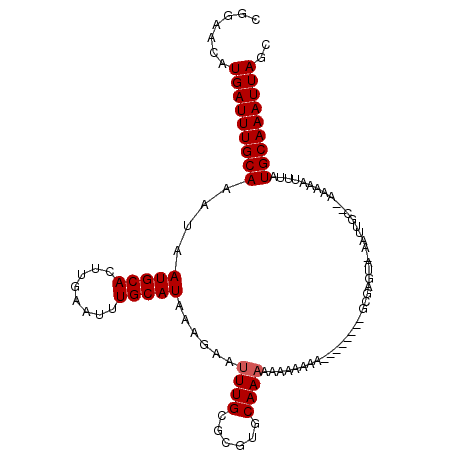
| Location | 10,282,709 – 10,282,814 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10282709 105 - 23771897 CGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAGAAUUUGCGAGUGCAAAAAAAAAAC---AAAAUGCGAGUAAAUUCGC--AAAAAUUUAUGCAAAUUAGC .......(((((((((.....((((((((.((((.......))))...))))))))..........---....((((((....)))))--)........))))))))).. ( -26.70) >DroVir_CAF1 189926 90 - 1 GGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAG--GUUUGC-----CAAAAAAAAAAA-AA-----------A-AACUGCCAAACAAUUUAUGCAAAUUAGC ............((((.....))))....(((((((((((.--(((((.-----((..........-..-----------.-...)).)))))...)))))))))))... ( -16.79) >DroPse_CAF1 170981 82 - 1 UGCAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAGAAUUUGC-----CAAAAAAAAAAA---G--------------------AAAAAAAUAUGCAAAUUAGC .......(((((((((....(((((........)))))......(((..-----....))).....---.--------------------.........))))))))).. ( -11.60) >DroSec_CAF1 157721 107 - 1 CGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAGAAUUUGCGCGUGCAAAAAAAAAAAAAAGAAAUGCGGGUU-UUUUGC--AAAAAUUUAUGCAAAUUAGC ............((((.....))))....((((((((((((...(((((((.((((.................)))).)).-....))--)))..))))))))))))... ( -22.53) >DroYak_CAF1 165888 101 - 1 CGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAGAAUUUGCGCGUGCAAAUAAAAAAA-------AGCGAGUAAACUCGC--AAAAAUUUAUGCAAAUUAGC ............((((.....))))....((((((((((((..((((((....)))))).......-------.(((((....)))))--.....))))))))))))... ( -27.60) >DroAna_CAF1 169766 99 - 1 CGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAAAAUUUGCACAACCAACAAAAAA---------CUCCAACGAAAUUGC--AAAAAUUUAUGCAAAUUAGC ............((((.....))))....((((((((((((...((((((..............---------............)))--)))..))))))))))))... ( -17.07) >consensus CGGAACAUGAUUUGCAAAUAAUGCACUUGAAUUUGCAUAAAGAAUUUGCGCGUGCAAAAAAAAAAA________GCGAGUA_AAUUGC__AAAAAUUUAUGCAAAUUAGC .......(((((((((....(((((........)))))......((((......)))).........................................))))))))).. (-13.30 = -13.47 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:45 2006