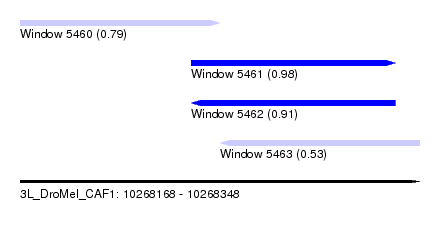
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,268,168 – 10,268,348 |
| Length | 180 |
| Max. P | 0.984670 |

| Location | 10,268,168 – 10,268,258 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -12.83 |
| Energy contribution | -12.55 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
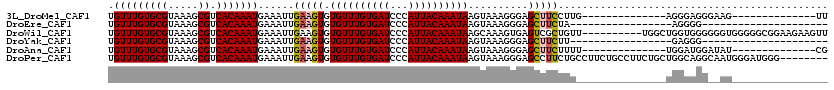
>3L_DroMel_CAF1 10268168 90 + 23771897 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAGAGUCCCC-GCA--UCCCCAAGAACUUCCCUCCCU ........(((......(((((....))))).((((((.............))))))...)))...-...--..................... ( -13.02) >DroSec_CAF1 143360 72 + 1 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAG---------------------AACUUCCCUCCUU ........((..((((.(((((....))))).((((((.............))))))))---------------------))..))....... ( -12.92) >DroSim_CAF1 149218 93 + 1 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAGAGUACCCCCCAGCCCCCCAAGAACUUCCCUCCUU ..........((((((.(((((....))))).((((((.............)))))))))))).............................. ( -13.62) >DroEre_CAF1 153545 71 + 1 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAGAGUCGCC---------------------CCCCU- ........(((......(((((....))))).((((((.............))))))...)))...---------------------.....- ( -13.92) >DroYak_CAF1 150931 71 + 1 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAGAGUCUCC---------------------CCCUC- .......((((......(((((....))))).((((((.............))))))...))))..---------------------.....- ( -14.02) >DroAna_CAF1 155907 92 + 1 UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCGCAUGCAAAUCCAUCAAAUGUGCUGCUGCUACUGCCGCCACCC-GCCGAUAUCCAUCCA ........((((((...(((((....)))))(((((((.............)))))))((.((....)).)).....-...))))))...... ( -21.62) >consensus UGUAAUAAGAUAUUUUAGUGACUUGUGUCACAGCACAUGCAAAUCCAUCAAAUGUGUAGAGUCCCC______________AACUUCCCUCCCU .................(((((....))))).((((((.............)))))).................................... (-12.83 = -12.55 + -0.28)

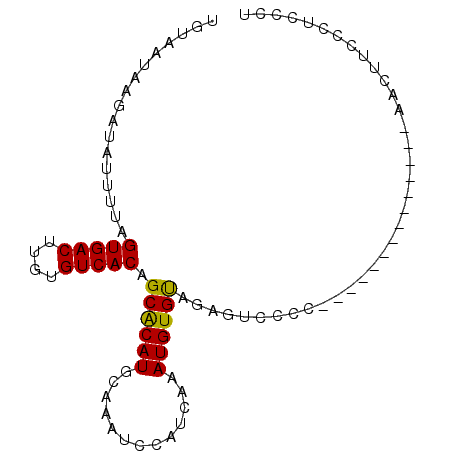

| Location | 10,268,245 – 10,268,337 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.79 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10268245 92 + 23771897 AA--------------CUUCCCUCCCU--------------CAAGGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA ..--------------(((((......--------------...))))).((((.((((......)))).))))......................((((((.((.....)))))))).. ( -19.30) >DroEre_CAF1 153611 82 + 1 ---------------------CCCCU-----------------UAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA ---------------------.....-----------------..(((((((((.((((......)))).))))(((((((...............))))))).)))))........... ( -18.26) >DroWil_CAF1 229796 110 + 1 AACUUCUUCCGCCCCCACCCCCCACCAGCCA----------AACAGCGACUCACUUUGCUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA ...........................((..----------...((((.((((..((((......)))))))).(((((((...............)))))))))))....))....... ( -15.16) >DroYak_CAF1 150997 82 + 1 ---------------------CCCUC-----------------AAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA ---------------------.....-----------------..(((((((((.((((......)))).))))(((((((...............))))))).)))))........... ( -18.26) >DroAna_CAF1 155986 92 + 1 CG--------------AUAUCCAUCCA--------------AAAAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA ..--------------...........--------------....(((((((((.((((......)))).))))(((((((...............))))))).)))))........... ( -18.26) >DroPer_CAF1 156712 112 + 1 --------CCCAUCCCAUUGCCUGCCAGCAGAAGGCAGAAGGCAGAAGGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA --------...(((((((((((((((.......)))))(((...(((((...))))).)))....)))))))))).....................((((((.((.....)))))))).. ( -30.90) >consensus ________________AU__CCCCCCA_______________AAAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACA .............................................(((((((((.((((......)))).))))(((((((...............))))))).)))))........... (-15.79 = -15.79 + 0.00)
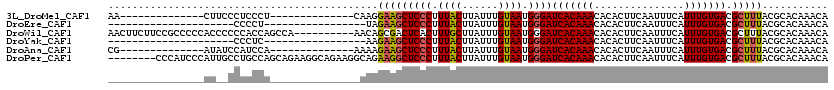
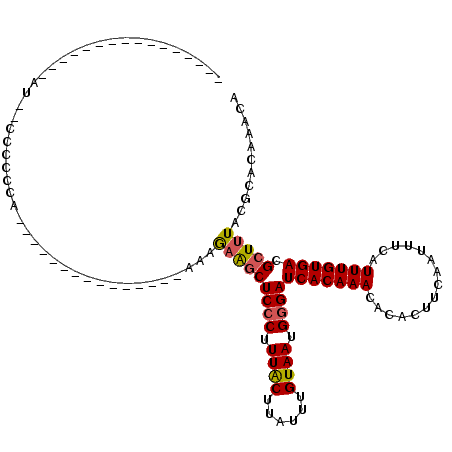
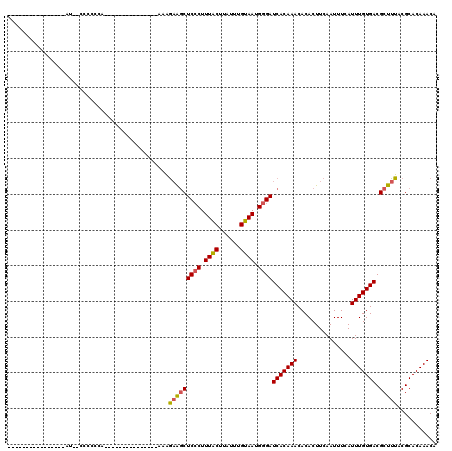
| Location | 10,268,245 – 10,268,337 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10268245 92 - 23771897 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUG--------------AGGGAGGGAAG--------------UU .(((((((((.....)).))))))).....................((((.((((......)))).))))((((((((.--------------....)))))))--------------). ( -26.50) >DroEre_CAF1 153611 82 - 1 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUA-----------------AGGGG--------------------- .(((((((((.....)).)))))))......(((((..........((((.((((......)))).)))))))))..-----------------.....--------------------- ( -20.20) >DroWil_CAF1 229796 110 - 1 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGCAAAGUGAGUCGCUGUU----------UGGCUGGUGGGGGGUGGGGGCGGAAGAAGUU .(((((((((.....)).)))))))..............(((.((..(((((..(..((.(((..((((...))))...----------..))).))..)..)))))..)).)))..... ( -27.60) >DroYak_CAF1 150997 82 - 1 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUU-----------------GAGGG--------------------- ........((..((((.((((((((.............))))))))((((.((((......)))).))))))))..)-----------------)....--------------------- ( -20.42) >DroAna_CAF1 155986 92 - 1 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUUUU--------------UGGAUGGAUAU--------------CG ((((..(.((..((((.((((((((.............))))))))((((.((((......)))).)))))))).....--------------)).)..)))).--------------.. ( -23.22) >DroPer_CAF1 156712 112 - 1 UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUUCUGCCUUCUGCCUUCUGCUGGCAGGCAAUGGGAUGGG-------- .(((((((((.....)).))))))).....................((((.((((......)))).)))).(((..(((((...(((.......))))))))..))).....-------- ( -32.50) >consensus UGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUUU_______________UGGGGGG__AU________________ .(((((((((.....)).)))))))......(((((.((((((((((...))))))))))..........)))))............................................. (-15.86 = -16.08 + 0.22)
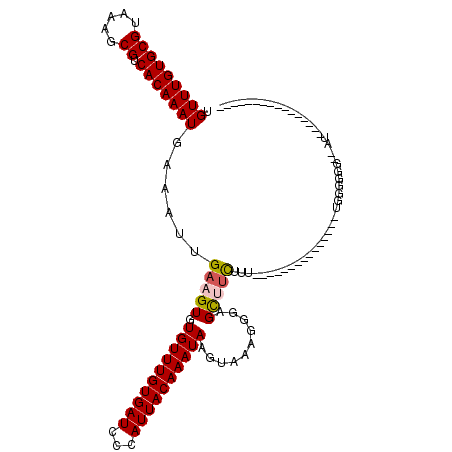
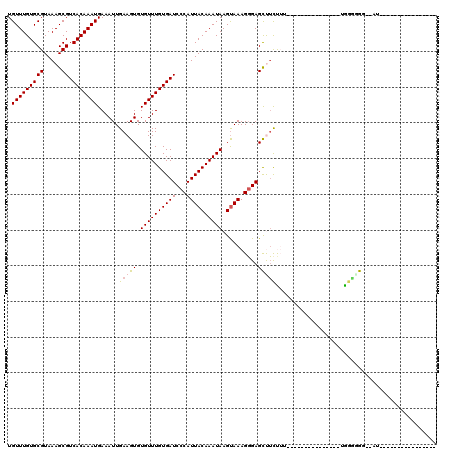
| Location | 10,268,258 – 10,268,348 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10268258 90 - 23771897 -------------------------AG----UCAGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUG- -------------------------((----..(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))..))..- ( -21.20) >DroPse_CAF1 156580 95 - 1 -----------------------AAAGCU--CUGACAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUUCUGCC -----------------------...(((--((..((((..(((((((((.....)).)))))))...)))).....((((((((((...)))))))))).......)))))........ ( -24.50) >DroGri_CAF1 156076 88 - 1 -----------------------ACUG-U--CUGUUGGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGCGCGC-G--CUGCGU--- -----------------------....-.--..((.(((((((((((..((((.(.(((((((((.............))))))))).)..))))..))))))))))-.--).))..--- ( -23.22) >DroWil_CAF1 229827 96 - 1 -----------------------UGUGCCCAGUGUCUGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGCAAAGUGAGUCGCUGUU- -----------------------......(((((.((....(((((((((.....)).))))))).........((.((((((((((...)))))))))).))......)).)))))..- ( -24.80) >DroYak_CAF1 151002 113 - 1 ACCAUCACCCCCCCCUUCCCACAAAAG----UUGGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUU--- ...................((((((..----....((((..(((((((((.....)).)))))))...)))).......)))))).((((.((((......)))).)))).......--- ( -26.52) >DroPer_CAF1 156744 95 - 1 -----------------------AAAGCU--CUGACAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUUCUGCC -----------------------...(((--((..((((..(((((((((.....)).)))))))...)))).....((((((((((...)))))))))).......)))))........ ( -24.50) >consensus _______________________AAAG_U__CUGACAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUCCU___ ...................................((((..(((((((((.....)).)))))))...))))..(..((((((((((...))))))))))..)................. (-16.62 = -16.32 + -0.30)

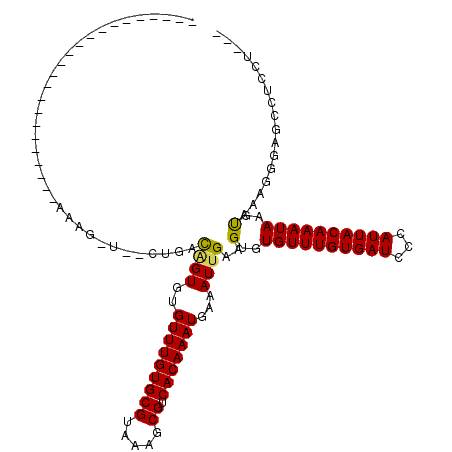
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:36 2006