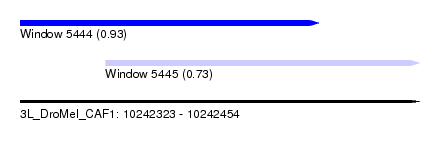
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,242,323 – 10,242,454 |
| Length | 131 |
| Max. P | 0.925734 |

| Location | 10,242,323 – 10,242,421 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -18.53 |
| Consensus MFE | -15.73 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
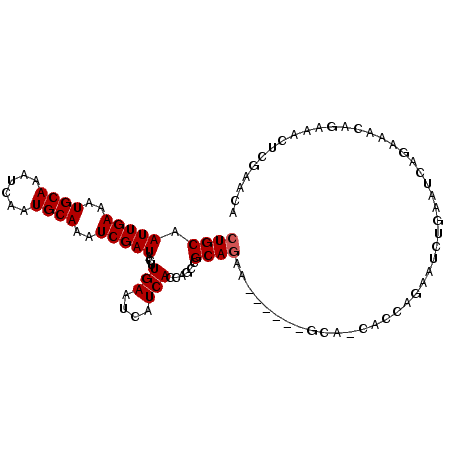
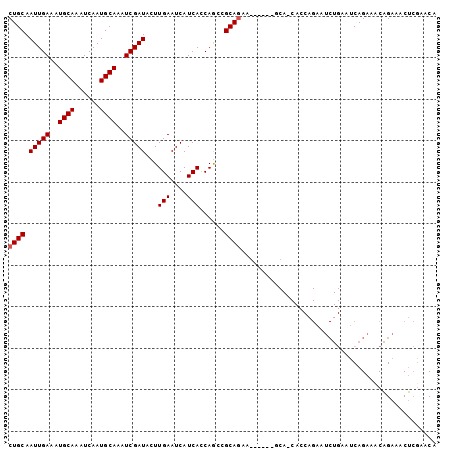
>3L_DroMel_CAF1 10242323 98 + 23771897 GCAUAAUCCAUUGUAAAGUGUCGAAAAGCUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAGCCGCAACAGCA---GCAGCACCA ((.............(((((((((.....((((.((((........))))))))..)))))))))............((.((....)).---)).)).... ( -21.80) >DroVir_CAF1 150956 87 + 1 AAAUAAUCCAUUGUAAAGUGUCGAAAAAGUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCAUCAACAGCAACAG-------------- ..........((((.(((((((((.....((((.((((........))))))))..)))))))))((....)).......))))...-------------- ( -16.80) >DroGri_CAF1 130339 87 + 1 AAAUAAUCCAUUGUAAAGUGUCGAAAAAGUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCAUGAACAGCAACAA-------------- ..........((((.(((((((((.....((((.((((........))))))))..)))))))))...((.....))...))))...-------------- ( -17.20) >DroSec_CAF1 118168 92 + 1 GCAUAAUCCAUUGUAAAGUGUCGAAAAGCUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAGCCGCAGAA---------GCAGCA (((........))).(((((((((.....((((.((((........))))))))..)))))))))............((.((....---------)).)). ( -19.70) >DroYak_CAF1 124530 92 + 1 GCAUAAUCCAUUGUAAAGUGUCGAAAAGCUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACCUGAAUCAUCACCAACCGCAGCGGCAGCA--------- ((......((((....)))).......(((((.(((((..((((......))))..)))))...(((....)))......)))))....)).--------- ( -19.20) >DroMoj_CAF1 170872 87 + 1 AAAUAAUCCAUUGUAAAGUGUCGAAAAAGUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCAUCAACAGCAGCAA-------------- ..........((((.(((((((((.....((((.((((........))))))))..)))))))))((....)).......))))...-------------- ( -16.50) >consensus AAAUAAUCCAUUGUAAAGUGUCGAAAAACUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAACAGCAACAG______________ ..........((((.(((((((((.....((((.((((........))))))))..)))))))))((....)).......))))................. (-15.73 = -15.65 + -0.08)



| Location | 10,242,351 – 10,242,454 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.28 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10242351 103 + 23771897 CUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAGCCGCAACAGCAGCAGCACCACCAGAAUCUGAAUCAGAAACAGAAACUCGAACA .(((.(((((..((((......))))..)))))................((.((....)).)).))).........((((.........)))).......... ( -15.40) >DroPse_CAF1 129318 78 + 1 CUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCAUCAAG-GCAGAG------------------------GCAGAGGCCGAGGCGGCAGCA ((((.(((((..((((......))))..))))).(((((.......)))))-((...(------------------------((....)))...)).)))).. ( -22.60) >DroSec_CAF1 118196 97 + 1 CUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAGCCGCAGAA------GCAGCACCAGAAUCUGAAUCAGAAUCAGAAACUCGAACA .((((.((((........))))))))..((((.................((.((....------)).)).......(((((.......)))))...))))... ( -17.00) >consensus CUGCAAUUGAAAUGCAAAUCAAUGCAAAUCGAUACUUGAAUCAUCACCAGCCGCAGAA______GCA_CACCAGAAUCUGAAUCAGAAACAGAAACUCGAACA ((((.(((((..((((......))))..)))))...(((....)))......))))............................................... (-10.27 = -10.60 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:19 2006