
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,232,223 – 10,232,314 |
| Length | 91 |
| Max. P | 0.984452 |

| Location | 10,232,223 – 10,232,314 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -15.76 |
| Energy contribution | -18.27 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10232223 91 + 23771897 --------------CUAGCAAGAAAGCCAUAGUAAGCUCAAAAUAAACCCCCUUCGUCCACGCAGGCAGCGCAAAAGUUGUCUGCGAUGGCAAACAGGGAGGAAA --------------..........(((........))).........((((((....(((((((((((((......)))))))))).))).....)))).))... ( -30.10) >DroGri_CAF1 121278 93 + 1 GGCUGCUUACGGCUGUAGAAAGAAAGCCAAAGUAAGCAUGAAAUAAACCCUCAUCGUCUACG---GCAGC---AGCGUUGUCUGCAGU-GCAAGCA-----AAAA .((((((....((((((((......((........))((((.........))))..))))))---)))))---))).(((..(((...-)))..))-----)... ( -26.30) >DroSec_CAF1 108141 91 + 1 --------------CUAGCAAGAAAGCCAAAGUAAGCUCGAAAUAAACCCCCAUCGUCCACGCAGGCAGCGCAAACGUUGUCUGCGAUUGCAAACAGGGAGGAAU --------------..........(((........))).........(((((........(((((((((((....)))))))))))..((....))))).))... ( -25.90) >DroSim_CAF1 113519 91 + 1 --------------CUAGCAAGAAAGCCAAAGUAAGCUCGAAAUAAACCCCCAUCGUCCACGCAGGCAGCGCAAACGUUGUCUGCGAUGGCAAACAGGGAGGAAU --------------..........(((........))).........(((((.....((((((((((((((....))))))))))).)))......))).))... ( -29.60) >DroEre_CAF1 116384 88 + 1 --------------CUGGCAAGAAAGCCAAAGUAAGCUCGAAAUAAACCCCCAGCGUCCACGCAGGCGAUGU---UGUUGUCUGCGAUGGCAAACAGGGAGGAAA --------------.((((......))))..................(((((.(..((((((((((((((..---.)))))))))).))).)..).))).))... ( -28.90) >DroYak_CAF1 114214 81 + 1 --------------UGGGCAAGAAAGCCAAAGUAAGCUCGAAAUAAACCCCCAUCGGCCACGCAA----------UGUUGUCUGCGAUGGCAAACGGGGAGGAAA --------------..(((......)))...................(((((....((((((((.----------.......)))).)))).....))).))... ( -24.80) >consensus ______________CUAGCAAGAAAGCCAAAGUAAGCUCGAAAUAAACCCCCAUCGUCCACGCAGGCAGCGC_AACGUUGUCUGCGAUGGCAAACAGGGAGGAAA .........................((........))..........(((((.....(((((((((((((......)))))))))).)))......))).))... (-15.76 = -18.27 + 2.50)

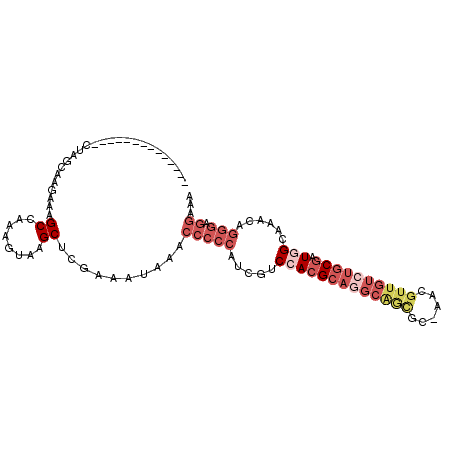

| Location | 10,232,223 – 10,232,314 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -14.98 |
| Energy contribution | -17.45 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10232223 91 - 23771897 UUUCCUCCCUGUUUGCCAUCGCAGACAACUUUUGCGCUGCCUGCGUGGACGAAGGGGGUUUAUUUUGAGCUUACUAUGGCUUUCUUGCUAG-------------- ....((((((((((((....))))))..(.((..(((.....)))..)).).))))))........(((((......))))).........-------------- ( -27.70) >DroGri_CAF1 121278 93 - 1 UUUU-----UGCUUGC-ACUGCAGACAACGCU---GCUGC---CGUAGACGAUGAGGGUUUAUUUCAUGCUUACUUUGGCUUUCUUUCUACAGCCGUAAGCAGCC ...(-----(((((((-...((((......))---)).((---.(((((..((((((.....))))))(((......)))......))))).)).)))))))).. ( -27.00) >DroSec_CAF1 108141 91 - 1 AUUCCUCCCUGUUUGCAAUCGCAGACAACGUUUGCGCUGCCUGCGUGGACGAUGGGGGUUUAUUUCGAGCUUACUUUGGCUUUCUUGCUAG-------------- ....((((((((((((....))))))).((((..(((.....)))..))))..)))))........(((((......))))).........-------------- ( -33.50) >DroSim_CAF1 113519 91 - 1 AUUCCUCCCUGUUUGCCAUCGCAGACAACGUUUGCGCUGCCUGCGUGGACGAUGGGGGUUUAUUUCGAGCUUACUUUGGCUUUCUUGCUAG-------------- ....((((((((((((....))))))).((((..(((.....)))..))))..)))))........(((((......))))).........-------------- ( -32.90) >DroEre_CAF1 116384 88 - 1 UUUCCUCCCUGUUUGCCAUCGCAGACAACA---ACAUCGCCUGCGUGGACGCUGGGGGUUUAUUUCGAGCUUACUUUGGCUUUCUUGCCAG-------------- ....(((((.((...(((.(((((......---.......))))))))..)).)))))..................((((......)))).-------------- ( -25.72) >DroYak_CAF1 114214 81 - 1 UUUCCUCCCCGUUUGCCAUCGCAGACAACA----------UUGCGUGGCCGAUGGGGGUUUAUUUCGAGCUUACUUUGGCUUUCUUGCCCA-------------- .....((((((((.((((.(((((......----------))))))))).))))))))........(((((......))))).........-------------- ( -28.90) >consensus UUUCCUCCCUGUUUGCCAUCGCAGACAACGUU_GCGCUGCCUGCGUGGACGAUGGGGGUUUAUUUCGAGCUUACUUUGGCUUUCUUGCUAG______________ .....((((((((..(((.(((((.((.(......).)).))))))))..))))))))........(((((......)))))....................... (-14.98 = -17.45 + 2.47)

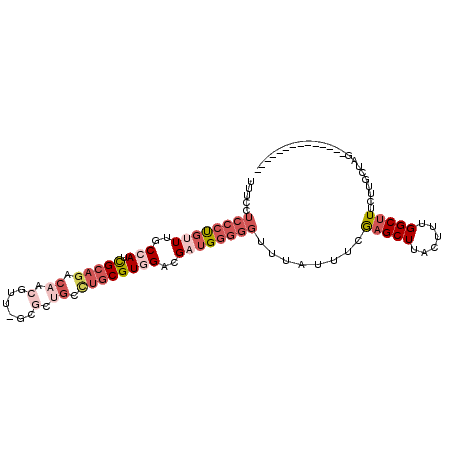
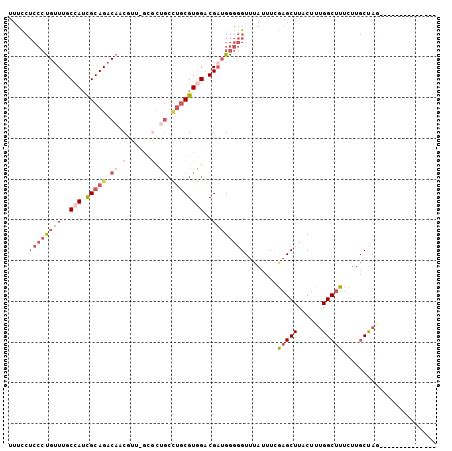
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:16 2006