
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,229,742 – 10,229,864 |
| Length | 122 |
| Max. P | 0.873905 |

| Location | 10,229,742 – 10,229,835 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -15.02 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
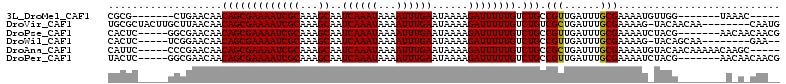
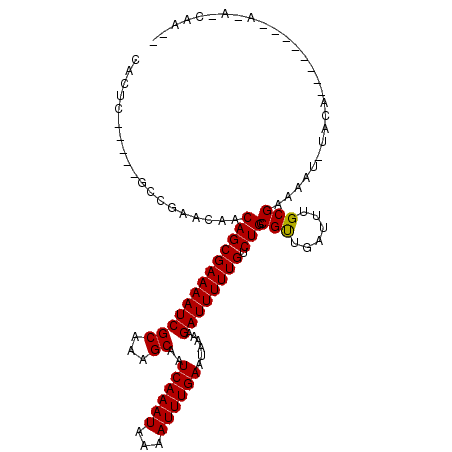
>3L_DroMel_CAF1 10229742 93 - 23771897 CGCG-------CUGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAAUGUUGG-------UAAAC----- ..((-------(((.....)))))....(((((((.(((((((((...)))))).....((((....))))....))).))))))).........-------.....----- ( -20.50) >DroVir_CAF1 135726 103 - 1 UGCGCUACUUGCUUAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGUCGCUGAUUUGCGAAAAG-UACAACAA--------CAAUG ...(.(((((.........(((((((((((((...))..((((((...))))))......)))))))).)))((((......)))).)))-))).....--------..... ( -20.70) >DroPse_CAF1 120253 100 - 1 CACUC-----GGCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAAUCUACG-------AACAACAACG ...((-----((((.((((..(((.....))).......((((((...))))))...........)))).))))((.(((((....))))).)))-------)......... ( -19.60) >DroWil_CAF1 172206 96 - 1 CACUC-----UCGGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAAG-UACAGCAA--------GAA-- ...((-----.(((.....(((((((((((((...))..((((((...))))))......)))))))).))))))..))(((((......-....))))--------)..-- ( -17.90) >DroAna_CAF1 109809 102 - 1 CAUUC-----CCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGCUGAUUUGCGAAAAUGUACAACAAAAACAAGC----- .....-----...(.(((.(((((((((((((...))..((((((...))))))......)))))))).))).(((......)))....))).).............----- ( -17.40) >DroPer_CAF1 120753 100 - 1 UACUC-----GGCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAAUCUACG-------AACAACAACG ...((-----((((.((((..(((.....))).......((((((...))))))...........)))).))))((.(((((....))))).)))-------)......... ( -19.60) >consensus CACUC_____GCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAAU_UACA________A_A_CAA__ ...................(((((((((((((...))..((((((...))))))......)))))))).))).(((......)))........................... (-15.02 = -14.80 + -0.22)
| Location | 10,229,762 – 10,229,864 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
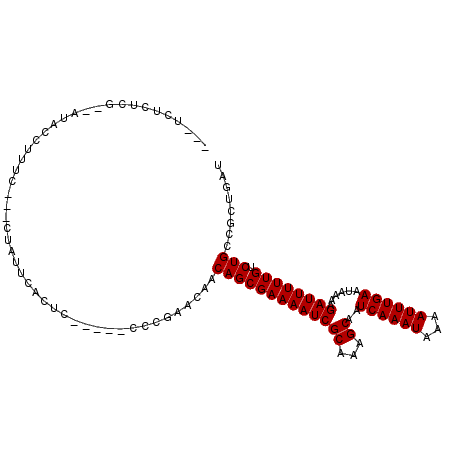
>3L_DroMel_CAF1 10229762 102 - 23771897 UCGUUUUUCGACAUACCUUUCUUCUUAUUCGCG-------CUGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAU .........((((......((((.((((((.((-------(((.....)))))......((...))...............))))))))))....)))).......... ( -17.40) >DroVir_CAF1 135749 105 - 1 ---UCGCUCGC-AGUAUUUCUCUGCUGCUUGCGCUACUUGCUUAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGUCGCUGAU ---..((..((-((((......))))))..))((.....))(((.(.((((((((((((((...))..((((((...))))))......)))))))).)))).).))). ( -26.20) >DroWil_CAF1 172227 96 - 1 GG-UGUGUC----UCCCCUUU---UCAUUCACUC-----UCGGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAU ((-..(((.----(((.....---..........-----..)))))).(((((((((((((...))..((((((...))))))......)))))))).)))))...... ( -15.03) >DroMoj_CAF1 154334 105 - 1 ---UCCCACGA-AGUCUUUCUGUGCUACUUGCUCGACGACAACGACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGUCGCUGAU ---........-.(((..((.(.((.....)).))).)))..(((((...(((((((((((...))..((((((...))))))......))))))))).)))))..... ( -20.70) >DroAna_CAF1 109836 96 - 1 -----UGUCAGCAUACCUUUC---CUAUUCAUUC-----CCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGCUGAU -----.((((((.........---...(((....-----...)))...(((((((((((((...))..((((((...))))))......)))))))).)))..)))))) ( -20.10) >DroPer_CAF1 120778 96 - 1 UC-UUUCUC----UACCUUUC---UUAUUUACUC-----GGCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAU ..-......----........---........((-----((((.....(((((((((((((...))..((((((...))))))......)))))))).))).)))))). ( -16.90) >consensus ___UCUCUCG__AUACCUUUC___CUAUUCACUC_____CCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGCUGAU ................................................(((((((((((((...))..((((((...))))))......)))))))).)))........ (-12.47 = -12.47 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:14 2006