
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,227,236 – 10,227,478 |
| Length | 242 |
| Max. P | 0.975492 |

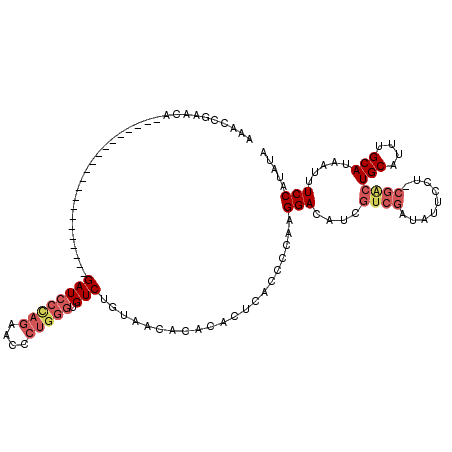
| Location | 10,227,236 – 10,227,334 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.98 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10227236 98 + 23771897 AAACCGAACA---------------------GAUCCCAGAACCCUGGGUGUCGGUAACACACACUCACCCCAAGGACAUCGUCGAUAUUCCU-CGACUGCAUUUGCAUAAUUUCCAUAUA ..(((((.((---------------------...(((((....))))))))))))..................(((....(((((......)-))))(((....))).....)))..... ( -25.20) >DroSec_CAF1 103121 98 + 1 AAACCGAACA---------------------GAUCCCAGAACCCUGGGUGUCUGUAACACACACUCACCCCAAGGACAUCGUCGAUAUUCCU-CGGCUGCAUUUGCAUAAUUUCCAUAUA .......(((---------------------((((((((....))))).))))))..................(((....(((((......)-))))(((....))).....)))..... ( -25.40) >DroSim_CAF1 108537 98 + 1 AAACCGAACA---------------------GAUCCCAGAACCCUGGGUGUCUGUAACACACACUCACCCCAAGGACAUCGUCGAUAUUCCU-CGACUGCAUUUGCAUAAUUUCCAUAUA .......(((---------------------((((((((....))))).))))))..................(((....(((((......)-))))(((....))).....)))..... ( -26.00) >DroEre_CAF1 109262 98 + 1 AAACCGAACA---------------------GAUCCCAGAUCCCUGGGUGUCUGUAACACACACUCACCCCAAGGACAUCGUCGAUAUUCCU-CGACUGCAUUUGCAUAAUUUCCAUAUA .......(((---------------------((((((((....))))).))))))..................(((....(((((......)-))))(((....))).....)))..... ( -26.00) >DroYak_CAF1 109022 119 + 1 AAACCGAACACAUCCCAUAUCCCAGAUACCAGAUCCUAGAUCCCUGGGUGUCUGUAACACACACUCACCCCAAGGACAUUGUCGAUAUGCCU-CGACUGCAUUUGCAUAAUUUCCAUAUA ......................((((((((.((((...))))....))))))))...................(((.((((((((......)-))))(((....))).))).)))..... ( -24.50) >DroAna_CAF1 107308 83 + 1 AAGCCGAAUA---------------------GAU-------CUCUGGGUGUCUGUGACCCCGUCU----CCCAGGAUAUA-----GAUUCCUGUGACUGCAUUUGCAUAAUUUCCAUAUA ..((.((((.---------------------...-------....(((.(((...))))))(((.----..(((((....-----...))))).)))...)))))).............. ( -18.00) >consensus AAACCGAACA_____________________GAUCCCAGAACCCUGGGUGUCUGUAACACACACUCACCCCAAGGACAUCGUCGAUAUUCCU_CGACUGCAUUUGCAUAAUUUCCAUAUA ...............................((((((((....))))).))).....................(((....((((.........))))(((....))).....)))..... (-11.70 = -12.98 + 1.28)


| Location | 10,227,295 – 10,227,385 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.12 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10227295 90 - 23771897 CUUAGAUCGAA-ACCCUUUCUUUGUAU-UCUCCUAACGGGGACGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGUCGAGGAAUAUCGAC ......((((.-..(((((((((((..-(((((....)))))..)))))))........((.....(((....))).))))))....)))). ( -24.60) >DroSec_CAF1 103180 89 - 1 CUUAGAUCGAA-ACCCUUUCUUUGUAU-UCUCCUAACGG-GACGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGCCGAGGAAUAUCGAC ......((((.-..(((((((((((..-((.((....))-))..))))))).......((......(((....))).))))))....)))). ( -21.40) >DroSim_CAF1 108596 89 - 1 CUUAGAUCGAA-ACCCUUUCUUUGUAU-UCUCCUAACGG-GACGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGUCGAGGAAUAUCGAC ......((((.-..(((((((((((..-((.((....))-))..)))))))........((.....(((....))).))))))....)))). ( -20.50) >DroEre_CAF1 109321 90 - 1 CUUAGAUCGAAAACCCUUUCUUUGUAU-UCUCCUAACGG-GGCGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGUCGAGGAAUAUCGAC ......((((....(((((((((((..-.((((....))-))..)))))))........((.....(((....))).))))))....)))). ( -23.10) >DroYak_CAF1 109102 90 - 1 CUUAGAUCGAA-ACCCUUUCUUUGUAUUUCUCCUAACGG-GGCGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGUCGAGGCAUAUCGAC ......((((.-..(((((((((((....((((....))-))..)))))))........((.....(((....))).))))))....)))). ( -22.00) >consensus CUUAGAUCGAA_ACCCUUUCUUUGUAU_UCUCCUAACGG_GACGACAGAGAAUUAUAUGGAAAUUAUGCAAAUGCAGUCGAGGAAUAUCGAC ......((((....(((((((((((...((.((....)).))..)))))))...............(((....)))...))))....)))). (-20.38 = -20.14 + -0.24)

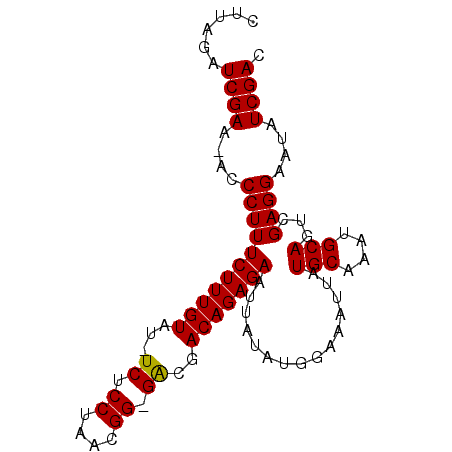

| Location | 10,227,385 – 10,227,478 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10227385 93 + 23771897 CGGUGAAUAUUUCUCUAUGCAAAUUUUCUGG------CGGCGUGUGAAAAUUCCAUGCCGCUCGCAGAGUCAAGACAUCUGCUCCUC---------------------GUUUUAUUUGCC .((..((((....................((------(((((((.((....))))))))))).(((((((....)).))))).....---------------------....))))..)) ( -30.00) >DroSec_CAF1 103269 93 + 1 CGGUGAAUAUUUCCCUAUGCAAAUUUUCUGG------CGGCGUGUGAAAACUCCAUGCCGCUCGCAGAGUCAAGACAUCUGCUCCUG---------------------GUUUUAUUUGCC .((..((((...((...............((------(((((((.((....))))))))))).(((((((....)).)))))....)---------------------)...))))..)) ( -32.70) >DroSim_CAF1 108685 93 + 1 CGGUGAAUAUUUCCCUAUGCAAAUUUUCUGG------CGGCGUGUGAAAAUUCCAUGCCGCUCGCAGAGUCAAGACAUCUGCUCCUG---------------------GUUUUAUUUGCC .((..((((...((...............((------(((((((.((....))))))))))).(((((((....)).)))))....)---------------------)...))))..)) ( -32.50) >DroEre_CAF1 109411 93 + 1 CGGUGAAUAUUUCCCUAUGCAAAUUUUCUGG------CGGCGUGUGAAAAUUCCAUGCCGCUUUCAGAGUCAAGACAUCUGCUCCUC---------------------GUUUUAUUUGCC .((..((((.........(((.....(((((------(((((((.((....))))))))))...))))((....))...))).....---------------------....))))..)) ( -27.27) >DroYak_CAF1 109192 93 + 1 CGGUGAAUAUUUUCCUAUGCAAAUUUUCUGG------CGGCGUGUGAAAAUACCAUGCCGCUUGCAGAGUCAAGACAUCUGCUCCUC---------------------GUUUUAUUUGCC .((..((((....................((------(((((((.(......)))))))))).(((((((....)).))))).....---------------------....))))..)) ( -28.30) >DroAna_CAF1 107467 117 + 1 --GAGAAUACUUCUCUAUGCAAAUUC-CUGGCCGGCGCGUCGUGUGAAAAUUUUAUGCCGCUUACAGAGUCAAGGCAGCUGCUCCUGCCAGGUUUUAGUUUUGUGUUUUUUUUUUUUGUC --((((((((....(((........(-(((((.((.(((.(..((....))....((((((((...))))...))))).))).)).))))))...)))....)))))))).......... ( -28.40) >consensus CGGUGAAUAUUUCCCUAUGCAAAUUUUCUGG______CGGCGUGUGAAAAUUCCAUGCCGCUCGCAGAGUCAAGACAUCUGCUCCUC_____________________GUUUUAUUUGCC .((..((((............................(((((((.((....)))))))))...(((((((....)).)))))..............................))))..)) (-17.85 = -18.47 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:12 2006