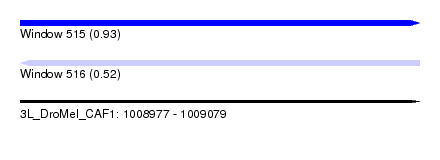
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,008,977 – 1,009,079 |
| Length | 102 |
| Max. P | 0.925765 |

| Location | 1,008,977 – 1,009,079 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.63 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
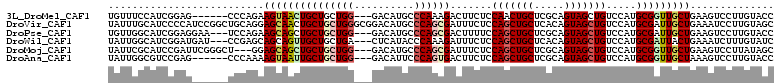
>3L_DroMel_CAF1 1008977 102 + 23771897 UGUUUCCAUCGGAG------CCCAGAAGUAACUGCUGCUGG---GACAUGCCCAAAGACUUCUCCAACUGCUCGCAGUAGCUGUCCAUGCGGUUGCUGAAGUCCUUGUACC ..........((..------(((((.(((....))).))))---).....))(((.((((((..(((((((..((((...))))....)))))))..)))))).))).... ( -34.70) >DroVir_CAF1 40594 111 + 1 UAUUUGCAUCCCCAUCCGGCUGCAGGAGCAACUGCUGCUGGGCGGACAUGCCCAGCGAUUUCUCCAGCUGCUCACAGUAGCUGUCCAUGCGAUUGCUGAAAUCCUUGUAGC ..................(((((((((((....)))((((((((....))))))))((((((..(((((((.....))))))).....((....)).)))))))))))))) ( -44.20) >DroPse_CAF1 38296 105 + 1 UGUUGGCAUCGGAGGAA---UCCAGAAGCAGCUGCUGCUGG---GACAUGCCCAGCGACUUUUCCAGCUGCUCGCAGUAGCUGUCCAUGCGAUUGCUGAAGUCCUUGUACC ....(((.((((...((---((((...((((((((((((((---(.....))))(((.((.....)).)))..)))))))))))...)).)))).)))).)))........ ( -37.70) >DroWil_CAF1 36643 105 + 1 UAUUGGCAUCGGAUGAU---CCGAGCAGCAGUUGCUGCUGA---CUCAUACCCAAAGAUUUCUCCAGCUGCUCACAGUAGCUGUCCAUGCGAUUACUGAAAUCUUUGUAUC ....((.(((....)))---))(((((((((...)))))).---))).....((((((((((..(((((((.....)))))))((.....)).....)))))))))).... ( -34.20) >DroMoj_CAF1 36387 105 + 1 UAUUCGCAUCCGAUUCGGGCU---GGAGCAGCUGCUGCUGG---GACAUGCCCAGCGAUUUCUCCAGCUGCUCGCAGUAGCUGUCCAUGCGGUUGCUGAAGUCCUUAUAGC ...(((....)))...(((((---.(((((((((..(((((---(.....))))))((....))))))))))).((((((((((....)))))))))).)))))....... ( -46.80) >DroAna_CAF1 32549 102 + 1 UAUUGGCGUCCGAG------CCCAAAAGUAAUUGCUGCUGG---GACAUUCCCAGUGACUUCUCCAGCUGCUCGCAGUAGCUGUCCAUGCGGUUGCUAAAGUCCUUGUACC ....(((......)------))....(((((((((.(((((---(.....))))))........(((((((.....))))))).....))))))))).............. ( -35.00) >consensus UAUUGGCAUCCGAG______CCCAGAAGCAACUGCUGCUGG___GACAUGCCCAGCGACUUCUCCAGCUGCUCGCAGUAGCUGUCCAUGCGAUUGCUGAAGUCCUUGUACC ..........................(((((((((((((((..........)))))).......(((((((.....))))))).....))))))))).............. (-22.83 = -23.63 + 0.81)
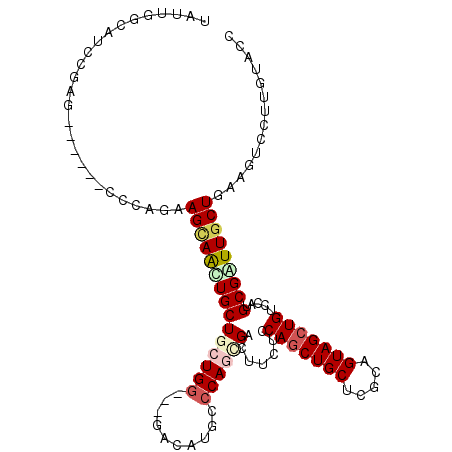
| Location | 1,008,977 – 1,009,079 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.43 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1008977 102 - 23771897 GGUACAAGGACUUCAGCAACCGCAUGGACAGCUACUGCGAGCAGUUGGAGAAGUCUUUGGGCAUGUC---CCAGCAGCAGUUACUUCUGGG------CUCCGAUGGAAACA .(..((((((((((..(((((((((((....))).))))....))))..))))))))))..)..(.(---((((.((......)).)))))------).....((....)) ( -33.90) >DroVir_CAF1 40594 111 - 1 GCUACAAGGAUUUCAGCAAUCGCAUGGACAGCUACUGUGAGCAGCUGGAGAAAUCGCUGGGCAUGUCCGCCCAGCAGCAGUUGCUCCUGCAGCCGGAUGGGGAUGCAAAUA ...............(((.((.(((...(.(((.(.(.(((((((((((....))(((((((......)))))))..)))))))))).).))).).))).)).)))..... ( -44.40) >DroPse_CAF1 38296 105 - 1 GGUACAAGGACUUCAGCAAUCGCAUGGACAGCUACUGCGAGCAGCUGGAAAAGUCGCUGGGCAUGUC---CCAGCAGCAGCUGCUUCUGGA---UUCCUCCGAUGCCAACA (((((.((((.(((((...((((((((....))).)))))(((((((((....))((((((.....)---)))))..)))))))..)))))---.))))..).)))).... ( -43.80) >DroWil_CAF1 36643 105 - 1 GAUACAAAGAUUUCAGUAAUCGCAUGGACAGCUACUGUGAGCAGCUGGAGAAAUCUUUGGGUAUGAG---UCAGCAGCAACUGCUGCUCGG---AUCAUCCGAUGCCAAUA ....(((((((((((((..((((((((....))).)))))...))))))...)))))))((((((((---.((((((...)))))))))((---.....)).))))).... ( -37.80) >DroMoj_CAF1 36387 105 - 1 GCUAUAAGGACUUCAGCAACCGCAUGGACAGCUACUGCGAGCAGCUGGAGAAAUCGCUGGGCAUGUC---CCAGCAGCAGCUGCUCC---AGCCCGAAUCGGAUGCGAAUA ...........(((.(((.(((..(((.(.((....))(((((((((((....))((((((.....)---)))))..))))))))).---.).)))...))).)))))).. ( -42.90) >DroAna_CAF1 32549 102 - 1 GGUACAAGGACUUUAGCAACCGCAUGGACAGCUACUGCGAGCAGCUGGAGAAGUCACUGGGAAUGUC---CCAGCAGCAAUUACUUUUGGG------CUCGGACGCCAAUA (((.((..(((((((((..((((((((....))).)))).)..)))...))))))..)).....(((---(.(((..(((......))).)------)).))))))).... ( -30.90) >consensus GGUACAAGGACUUCAGCAACCGCAUGGACAGCUACUGCGAGCAGCUGGAGAAAUCGCUGGGCAUGUC___CCAGCAGCAGCUGCUUCUGGA______CUCCGAUGCCAAUA ....((..((((((.((....)).....(((((.(.....).)))))..))))))..))...........((((.(((....))).))))..................... (-16.85 = -17.27 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:46 2006