
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
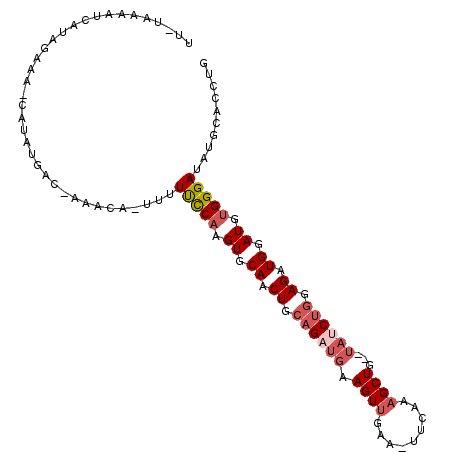
| Location | 10,221,761 – 10,221,865 |
| Length | 104 |
| Max. P | 0.564073 |

| Location | 10,221,761 – 10,221,865 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -15.23 |
| Energy contribution | -16.28 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10221761 104 + 23771897 UUAUAAAAUCAUAGAAAUCAUAUGAC-AAACA-UUUUUUCAAGUGCAACUGCAGUCGAAGUUGAA-UUCAAAGCUG--UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ........(((((.......))))).-...((-(..(..((.((.((.((.(((....((((...-.....)))).--...))).)).)).)).))..).)))...... ( -18.20) >DroSec_CAF1 97903 103 + 1 UUUAAAAAUCAUGGACA-CAUAUGAC-AUACA-UUUUCCCAAGUGCAACUGCAGAUGAAGUUGAA-UUCAAAGCUG--UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ........(((((....-..))))).-...((-(..(((((.((.((.((.((((((.((((...-.....)))).--)))))).)).)).)).))))).)))...... ( -27.70) >DroSim_CAF1 103252 103 + 1 UUAUAAAAUCAUAGAAA-CAUAUGAC-AAACA-UUUUUCCAAGUGCAACUGCAGAUGAAGUUGAA-UUCAAAGCUG--UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ........(((((....-..))))).-...((-(..(((((.((.((.((.((((((.((((...-.....)))).--)))))).)).)).)).))))).)))...... ( -25.30) >DroEre_CAF1 103580 103 + 1 AUGUAAAAUUAUAAAAG-CAUAUGAC-AAACA-UUUUUCCAAGUGCAGCUGCAGAUGAAGUUAAA-UUCAAAGCUG--UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ................(-(((((..(-(....-..((((((..(((((((.....((((......-)))).)))))--))..))))))......))..))))))..... ( -30.70) >DroYak_CAF1 103269 94 + 1 ---------AAAAGAAA-CAGAUGGC-AAACA-UUUUUCCAAGUGCAACUGCAGAUGAAGUUGAA-UUCAAAGCUG--UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ---------........-(((...((-(....-...(((((.((.((.((.((((((.((((...-.....)))).--)))))).)).)).)).)))))..)))..))) ( -24.40) >DroAna_CAF1 99427 96 + 1 ------------GCUAA-CUUAAGCCCAAUUAUUUUUUCCAAGUGCAUCUGCCGAUGAAGUCAGCCUGUGGAGCUGGGAACCUAGAGAUGGAUGCGGGAUAUGCACCUG ------------(((..-....)))..............((.((((((.(.(((......((((((....).)))))...((.......))...))).).)))))).)) ( -21.60) >consensus UU_UAAAAUCAUAGAAA_CAUAUGAC_AAACA_UUUUUCCAAGUGCAACUGCAGAUGAAGUUGAA_UUCAAAGCUG__UAUCUGGAGAUGGAUGUGGGAUAUGCACCUG ....................................(((((.((.((.((.((((((.((((.........))))...)))))).)).)).)).))))).......... (-15.23 = -16.28 + 1.06)


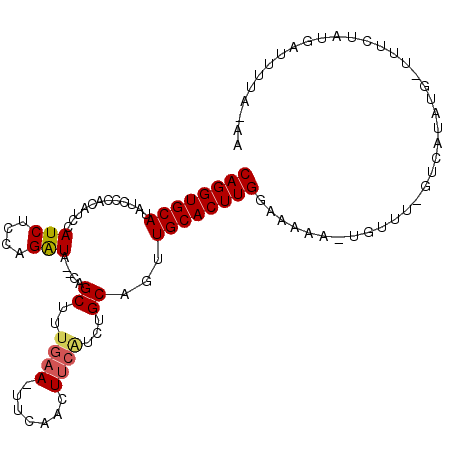
| Location | 10,221,761 – 10,221,865 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -13.22 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10221761 104 - 23771897 CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA--CAGCUUUGAA-UUCAACUUCGACUGCAGUUGCACUUGAAAAAA-UGUUU-GUCAUAUGAUUUCUAUGAUUUUAUAA ((((((((............(((....))).--..((.(((((-......)))))..))...))))))))......-(((..-((((((.(....)))))))...))). ( -19.40) >DroSec_CAF1 97903 103 - 1 CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA--CAGCUUUGAA-UUCAACUUCAUCUGCAGUUGCACUUGGGAAAA-UGUAU-GUCAUAUG-UGUCCAUGAUUUUUAAA ((((((((............(((....))).--..((..((((-......))))...))...))))))))(((..(-(((..-...)))).-..)))............ ( -19.80) >DroSim_CAF1 103252 103 - 1 CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA--CAGCUUUGAA-UUCAACUUCAUCUGCAGUUGCACUUGGAAAAA-UGUUU-GUCAUAUG-UUUCUAUGAUUUUAUAA ((((((((............(((....))).--..((..((((-......))))...))...))))))))......-(((..-((((((..-....))))))...))). ( -19.60) >DroEre_CAF1 103580 103 - 1 CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA--CAGCUUUGAA-UUUAACUUCAUCUGCAGCUGCACUUGGAAAAA-UGUUU-GUCAUAUG-CUUUUAUAAUUUUACAU .....((((((...(((..(((.(((((...--(((((.((((-......)))).....)))))...)))))...)-))..)-)).)))))-)................ ( -20.70) >DroYak_CAF1 103269 94 - 1 CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA--CAGCUUUGAA-UUCAACUUCAUCUGCAGUUGCACUUGGAAAAA-UGUUU-GCCAUCUG-UUUCUUUU--------- ((((((((............(((....))).--..((..((((-......))))...))...))))))))((((.(-((...-..)))...-))))....--------- ( -16.70) >DroAna_CAF1 99427 96 - 1 CAGGUGCAUAUCCCGCAUCCAUCUCUAGGUUCCCAGCUCCACAGGCUGACUUCAUCGGCAGAUGCACUUGGAAAAAAUAAUUGGGCUUAAG-UUAGC------------ (((((((((...(((...((.......))....(((((.....))))).......)))...)))))))))..............(((....-..)))------------ ( -23.20) >consensus CAGGUGCAUAUCCCACAUCCAUCUCCAGAUA__CAGCUUUGAA_UUCAACUUCAUCUGCAGUUGCACUUGGAAAAA_UGUUU_GUCAUAUG_UUUCUAUGAUUUUA_AA ((((((((............(((....))).....((..((((.......))))...))...))))))))....................................... (-13.22 = -13.47 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:04 2006