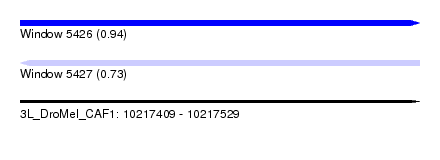
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,217,409 – 10,217,529 |
| Length | 120 |
| Max. P | 0.944767 |

| Location | 10,217,409 – 10,217,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -26.53 |
| Energy contribution | -25.40 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10217409 120 + 23771897 AAAGUCGGCGGUCUUAUUGAAGCUGGUUUCUGACUCCUCAUGAUUAGCAACUUCAACAUGGUGCAAUAGGAUUUGCUGCACUGAAACCACUUUUGGUUGUAAGCGGCAUCGCCCACUUUC (((((.(((((((((((((..(((((((..(((....))).))))))).(((.......))).))))))))).((((((.....(((((....)))))....)))))).)))).))))). ( -39.60) >DroPse_CAF1 107784 120 + 1 AAUGUGGGCGGCCUGAUCGAGGCCGGCCUCUGGCUGCGCAUGAUCAGCAUCUUGAGCAUUUGGCAGUAGGUUUUGCUGCACUGAAACCACUUCUGAUUGUAGGCAGCGUCGCCCACUCGC ...(((((((((......((((....))))..(((((..(..(((((........((.....))(((.((((((........))))))))).)))))..)..)))))))))))))).... ( -48.80) >DroGri_CAF1 107047 120 + 1 AAUGUGGGCGGUCUUAUCAUCGCUGGCUUUUGGCUGCGCAAAAUGAGCAUCUUAAGCAUGAUGCAAUAAGUUUUGCUGCACUGAAACCAGUUCUGAUUGUAGGCCGCAUCACCCACACGC ..((((((((((((.((((..(((((.(((..(.((((((((((..(((((........))))).....))))))).))))..))))))))..))))...))))))).....)))))... ( -49.50) >DroWil_CAF1 156014 120 + 1 AAUGUGGGUGGCUUUAUGGCAGCGGGUCUCUGGCUACGCAUAAUCAAUAACUUCAACAUUUUGCAAUAGGUUUUGCUGCAUUGAAACCAUUUUUGAUUAUACGCUGAAUCGCCCACUCGC ...(((((((..((...(((.((((((.....))).)))((((((((....(((((......((((......))))....))))).......))))))))..)))))..))))))).... ( -33.30) >DroMoj_CAF1 138211 120 + 1 AAAGUGGGUGGCCUUAUCAUCGCUGGCUUCUGGCUGCGCAUAAUCAUCAGCUUGAGCAUGAUGCAAUACCUCUUGCUGCACUGGAACCAGUUCUGGUUGUAGGCGGCAUCGCCCACGCGU ...((((((((......(((((...((((..(((((...........))))).)))).)))))..........((((((.(((.(((((....))))).))))))))))))))))).... ( -46.50) >DroPer_CAF1 108190 120 + 1 AAUGUGGGCGGCCUGAUCGAGGCCGGCCUCUGGCUGCGCAUGAUCAGCAUCUUGAGCAUUUGGCAGUAGGUUUUGCUGCACUGAAACCACCUCUGAUUGUAGGCAGCGUCGCCCACUCGC ...(((((((((......((((....))))..(((((..(..(((((.........((....(((((((...)))))))..)).........)))))..)..)))))))))))))).... ( -48.57) >consensus AAUGUGGGCGGCCUUAUCGAAGCUGGCCUCUGGCUGCGCAUAAUCAGCAUCUUGAGCAUGUUGCAAUAGGUUUUGCUGCACUGAAACCACUUCUGAUUGUAGGCAGCAUCGCCCACUCGC ...(((((((((.(..((...(((((((...))))).))((((((((.........((....((((......)))).....)).........)))))))).))..).))))))))).... (-26.53 = -25.40 + -1.13)


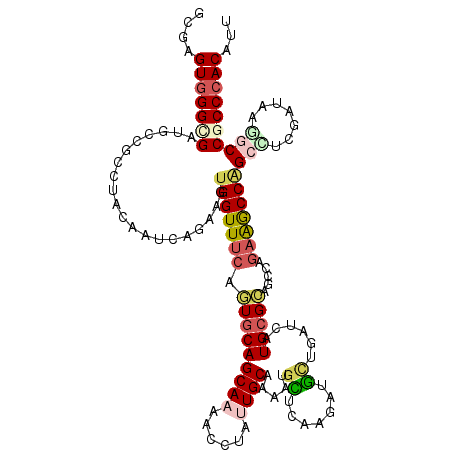
| Location | 10,217,409 – 10,217,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -22.22 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10217409 120 - 23771897 GAAAGUGGGCGAUGCCGCUUACAACCAAAAGUGGUUUCAGUGCAGCAAAUCCUAUUGCACCAUGUUGAAGUUGCUAAUCAUGAGGAGUCAGAAACCAGCUUCAAUAAGACCGCCGACUUU .(((((.(((((.(((((((........))))))).)).((((((.........))))))..(((((((((((....((.(((....)))))...))))))))))).....))).))))) ( -39.10) >DroPse_CAF1 107784 120 - 1 GCGAGUGGGCGACGCUGCCUACAAUCAGAAGUGGUUUCAGUGCAGCAAAACCUACUGCCAAAUGCUCAAGAUGCUGAUCAUGCGCAGCCAGAGGCCGGCCUCGAUCAGGCCGCCCACAUU ....((((((...(((((((..(((((....)))))..)).)))))....(((.(((.....(((.((.(((....))).)).)))..))))))..(((((.....)))))))))))... ( -45.00) >DroGri_CAF1 107047 120 - 1 GCGUGUGGGUGAUGCGGCCUACAAUCAGAACUGGUUUCAGUGCAGCAAAACUUAUUGCAUCAUGCUUAAGAUGCUCAUUUUGCGCAGCCAAAAGCCAGCGAUGAUAAGACCGCCCACAUU ..(((((((((....(.....).((((...(((((((..(.((.((((((......(((((........)))))...))))))...)))..)))))))...)))).....))))))))). ( -38.70) >DroWil_CAF1 156014 120 - 1 GCGAGUGGGCGAUUCAGCGUAUAAUCAAAAAUGGUUUCAAUGCAGCAAAACCUAUUGCAAAAUGUUGAAGUUAUUGAUUAUGCGUAGCCAGAGACCCGCUGCCAUAAAGCCACCCACAUU ....(((((.(.....(((((((((((...((..((((((((..((((......))))....))))))))..)))))))))))))............(((.......)))).)))))... ( -33.60) >DroMoj_CAF1 138211 120 - 1 ACGCGUGGGCGAUGCCGCCUACAACCAGAACUGGUUCCAGUGCAGCAAGAGGUAUUGCAUCAUGCUCAAGCUGAUGAUUAUGCGCAGCCAGAAGCCAGCGAUGAUAAGGCCACCCACUUU .((((((((((....)))))))........((((((...(((((((((......))))((((.((....)))))).....)))))))))))......))).......((....))..... ( -39.10) >DroPer_CAF1 108190 120 - 1 GCGAGUGGGCGACGCUGCCUACAAUCAGAGGUGGUUUCAGUGCAGCAAAACCUACUGCCAAAUGCUCAAGAUGCUGAUCAUGCGCAGCCAGAGGCCGGCCUCGAUCAGGCCGCCCACAUU ....(((((((.....((((...(((.(((((((((((.(.((((.........)))))...(((.((.(((....))).)).)))....)))))).)))))))).)))))))))))... ( -45.30) >consensus GCGAGUGGGCGAUGCCGCCUACAAUCAGAAGUGGUUUCAGUGCAGCAAAACCUAUUGCAAAAUGCUCAAGAUGCUGAUCAUGCGCAGCCAGAAGCCAGCCUCGAUAAGGCCGCCCACAUU ....(((((((....................(((((((.(((((((((......)))).....((.......))......))))).....)))))))(((.......))))))))))... (-22.22 = -23.12 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:02 2006