
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,217,149 – 10,217,239 |
| Length | 90 |
| Max. P | 0.958519 |

| Location | 10,217,149 – 10,217,239 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
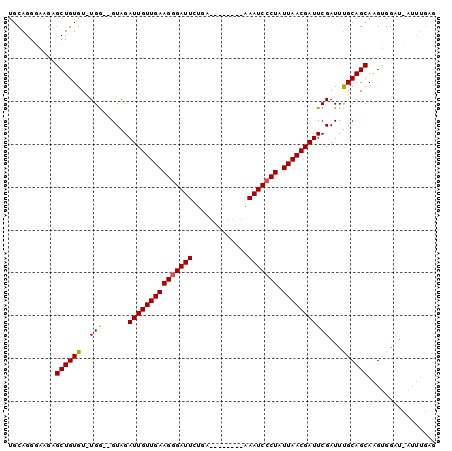
>3L_DroMel_CAF1 10217149 90 + 23771897 UGCAGGGAAGAGCUGUGUUUGG--GUAGAUUGUUGAAGGGAUUCUGA--------AAAUCCCUAUUAACGAUUCGAUUUGCAGCAAGUGGAUCAUUUGAG .((((.......))))..(..(--((.((((((((((((((((....--------.))))))).))))))))).((((..(.....)..)))))))..). ( -25.60) >DroSim_CAF1 97866 81 + 1 UGCAGGGAAGAGCUGUGU-UGA--GUAGAUUGUUGAAGGGAUUCUGA--------AAAUCCCUAUUAACGAUUCGAUUUGCAGCAAGCGGA--------A ...........(((((((-(((--....(((((((((((((((....--------.))))))).)))))))))))))..))))).......--------. ( -24.10) >DroYak_CAF1 98304 99 + 1 UGCAGGCAAGAGCUGUGU-UGGGUGUGGAUUGUUGAAGAGAUUCUGAUUAUCCCUGAAUCCCUAUUAACGAUUCGAUUCGCAGCAAGUGGCUGAUUUGAG ..(((.((...((((((.-......(((((((((((((.(((((...........))))).)).)))))))))))...))))))...)).)))....... ( -30.70) >consensus UGCAGGGAAGAGCUGUGU_UGG__GUAGAUUGUUGAAGGGAUUCUGA________AAAUCCCUAUUAACGAUUCGAUUUGCAGCAAGUGGAU_AUUUGAG ...........((((((..(((......(((((((((((((((.............))))))).)))))))))))...))))))................ (-18.83 = -18.72 + -0.11)


| Location | 10,217,149 – 10,217,239 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -14.77 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
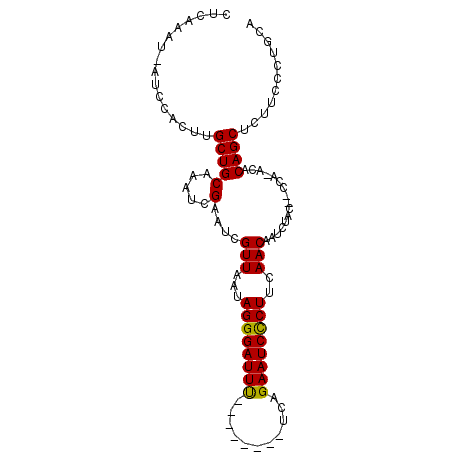
>3L_DroMel_CAF1 10217149 90 - 23771897 CUCAAAUGAUCCACUUGCUGCAAAUCGAAUCGUUAAUAGGGAUUU--------UCAGAAUCCCUUCAACAAUCUAC--CCAAACACAGCUCUUCCCUGCA ................(((((.....)....(((...((((((((--------...))))))))..))).......--.......))))........... ( -12.60) >DroSim_CAF1 97866 81 - 1 U--------UCCGCUUGCUGCAAAUCGAAUCGUUAAUAGGGAUUU--------UCAGAAUCCCUUCAACAAUCUAC--UCA-ACACAGCUCUUCCCUGCA .--------...((.....))..........(((...((((((((--------...))))))))..))).......--...-.....((........)). ( -13.80) >DroYak_CAF1 98304 99 - 1 CUCAAAUCAGCCACUUGCUGCGAAUCGAAUCGUUAAUAGGGAUUCAGGGAUAAUCAGAAUCUCUUCAACAAUCCACACCCA-ACACAGCUCUUGCCUGCA .......(((.((...((((.(....(((((.(.....).)))))((((((.......)))))).................-.).))))...)).))).. ( -17.90) >consensus CUCAAAU_AUCCACUUGCUGCAAAUCGAAUCGUUAAUAGGGAUUU________UCAGAAUCCCUUCAACAAUCUAC__CCA_ACACAGCUCUUCCCUGCA ................(((((.....)....(((...((((((((...........))))))))..)))................))))........... (-12.67 = -12.23 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:00 2006