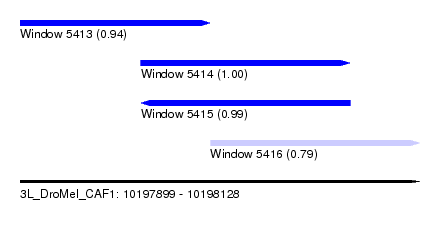
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,197,899 – 10,198,128 |
| Length | 229 |
| Max. P | 0.995017 |

| Location | 10,197,899 – 10,198,008 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -55.90 |
| Consensus MFE | -38.40 |
| Energy contribution | -41.32 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10197899 109 + 23771897 CGGUGGAUAACCGCCGAAUGCGUUUGGUGGUCCUCCUGGUCCUGCUGGUCCGGGAGGAUAUCCGCCGAAGGCCUGGGGCGGUGGUGCUCCUCCUCCGUAGUGUGGCUGU (((((((((.((((((((....))))))))(((((((((.((....)).))))))))))))))))))..((((....((((.((.......)).)))).....)))).. ( -60.90) >DroSec_CAF1 74288 109 + 1 CGGUGGAUAACCGGCGAAUGCGUUUGGUGGUCCCCCUGGUCCUGCUGGUCCGGGAGGAUAUCCGCCGAAGGCAUGUGGUGGUGGUGCUCCUCCUCCGUAGGGCGGCUGU .((.(((..((((.((.((((.(((((((((((.(((((.((....)).))))).)))...)))))))).))))....)).))))..))).)).(((.....))).... ( -51.90) >DroSim_CAF1 78454 109 + 1 CGGUGGAUAACCGCCGAAUGCGUUUGGUGGUCCCCCUGGUCCUGCUGGUCCGGGAGGAUAUCCGCCGAAGGCAUGUGGUGGUGGUGCUCCUCCUCCGUAGGGCGGCUGU .((.(((..(((((((.((((.(((((((((((.(((((.((....)).))))).)))...)))))))).))))....)))))))..))).)).(((.....))).... ( -56.70) >DroEre_CAF1 78970 109 + 1 UGGUGGAUACCCGCCGAAUGCGUUGGCAGGUCCUCCUGGUCCUGCUGGUCCGGGGGGAUAACCGCCGAAGGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGGUGGCUGU .(((((...((.(((((.....))))).))(((((((((.((....)).)))))))))...)))))(..(.((((((((((.((....)).)))))))))).)..)... ( -57.30) >DroYak_CAF1 78355 100 + 1 CGGUGGAUAUCCGCCAAAUGCGCUUGGAGGAC---------CUGCUGGUCCUGGGGGAUACCCUCCAAAGGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGCUGGCUGU .(((((....)))))....((..(((((((((---------(....))))))((((....)))))))).((((((((((((.((....)).)))))))))))).))... ( -52.70) >consensus CGGUGGAUAACCGCCGAAUGCGUUUGGUGGUCCCCCUGGUCCUGCUGGUCCGGGAGGAUAUCCGCCGAAGGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGGUGGCUGU ((((((....))))))...((.(((((((((((.(((((.((....)).))))).)))...)))))))).))((((((.((.((....)).)).))))))......... (-38.40 = -41.32 + 2.92)



| Location | 10,197,968 – 10,198,088 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -55.84 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.44 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10197968 120 + 23771897 GGCCUGGGGCGGUGGUGCUCCUCCUCCGUAGUGUGGCUGUCCACCAGGACCACCUACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUA .((.((((((((((((.(.........((((.((((...(((....)))))))))))...((((((....))))))).)))).))))...((((((....))))))......)))).)). ( -51.90) >DroSec_CAF1 74357 120 + 1 GGCAUGUGGUGGUGGUGCUCCUCCUCCGUAGGGCGGCUGUCCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGACCACCCAUGUA .(((((.((((((((((.((..((((((((..((((..(((((...((((...((((.....))))..))))...)))))...)))).))))))))..)).))))...))))))))))). ( -64.10) >DroSim_CAF1 78523 120 + 1 GGCAUGUGGUGGUGGUGCUCCUCCUCCGUAGGGCGGCUGUCCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGUCCACCCAUGUA .(((((.(((((.((((.((..((((((((..((((..(((((...((((...((((.....))))..))))...)))))...)))).))))))))..)).))))....)))))))))). ( -63.70) >DroEre_CAF1 79039 108 + 1 GGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGGUGGCUGUCCACCGGGACCACCUCCAUAAGGUG------------GAGCACCCGCAUAAGGUGGCUGGCCACCUGGACCACCCACGUA ..((((((((((.((....)).))))))))))((((..((((..((((.((((((.....)))))------------)....))))....((((((....))))))))))...))))... ( -54.90) >DroYak_CAF1 78415 90 + 1 GGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGCUGGCUGUCCACCUGGACCA------------------------------CCCACAUAAGGUGGCUGUCCACCUGGACCACCCACGUA ((((((((((((.((....)).))))))))))))((..(((((..(((((..------------------------------.((((.....))))..)))))..)))))..))...... ( -44.60) >consensus GGCCUGUGGAGGUGGUGCUCCUCCUCCGUAGGGUGGCUGUCCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUA .((.((.((.(((((((.((..((.((.....((((..((((....))))((((.......))))..................))))....)).))..)).))))...))).)))).)). (-29.86 = -30.44 + 0.58)



| Location | 10,197,968 – 10,198,088 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -56.36 |
| Consensus MFE | -36.25 |
| Energy contribution | -38.88 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10197968 120 - 23771897 UACAUGGGUGGUCCAGGUGGUCAGCCACCUUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUAGGUGGUCCUGGUGGACAGCCACACUACGGAGGAGGAGCACCACCGCCCCAGGCC .....(((((((..((((((....))))))......((((.(((.(((.....))).((((.((((((((((((....))...))))).))))))))).))).)))))))))))...... ( -57.70) >DroSec_CAF1 74357 120 - 1 UACAUGGGUGGUCCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGGUCCUGGUGGACAGCCGCCCUACGGAGGAGGAGCACCACCACCACAUGCC ..((((((((((...((((.((..((.(((((.((((.((..(((((......(((((((.....))))))))))))....)).))))..))))).))..)).)))))))))).)))).. ( -63.60) >DroSim_CAF1 78523 120 - 1 UACAUGGGUGGACCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGGUCCUGGUGGACAGCCGCCCUACGGAGGAGGAGCACCACCACCACAUGCC ..(((((((((....((((.((..((.(((((.((((.((..(((((......(((((((.....))))))))))))....)).))))..))))).))..)).)))).))))).)))).. ( -61.80) >DroEre_CAF1 79039 108 - 1 UACGUGGGUGGUCCAGGUGGCCAGCCACCUUAUGCGGGUGCUC------------CACCUUAUGGAGGUGGUCCCGGUGGACAGCCACCCUACGGAGGAGGAGCACCACCUCCACAGGCC ...(((((.(((..((((((....))))))......(((((((------------(.((((.(((.((((((((....))...)))))))))..)))).))))))))))))))))..... ( -54.40) >DroYak_CAF1 78415 90 - 1 UACGUGGGUGGUCCAGGUGGACAGCCACCUUAUGUGGG------------------------------UGGUCCAGGUGGACAGCCAGCCUACGGAGGAGGAGCACCACCUCCACAGGCC ......(((.(((((..(((...(((((((.....)))------------------------------)))))))..))))).))).((((..(((((.((....)).)))))..)))). ( -44.30) >consensus UACAUGGGUGGUCCAGGUGGUCAGCCACCUUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUAGGUGGUCCUGGUGGACAGCCACCCUACGGAGGAGGAGCACCACCACCACAGGCC .......((((....((((((...((.((((....(((((((.............(((((.....)))))((((....)))).)))))))....)))).))...)))))).))))..... (-36.25 = -38.88 + 2.63)


| Location | 10,198,008 – 10,198,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -47.58 |
| Consensus MFE | -32.83 |
| Energy contribution | -34.94 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10198008 120 + 23771897 CCACCAGGACCACCUACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAU ...((.(((((((((.....))))))...)))(((.((((..........((((((....)))))).(((((((.......))))))))))))))......))((((((...)))))).. ( -51.80) >DroSec_CAF1 74397 120 + 1 CCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAC ...(((((..((.((((.....)))).))....)))))..........(((((((((.((((((((..((.......)).))))))))))))(((((((.....)))))))...))))). ( -51.70) >DroSim_CAF1 78563 120 + 1 CCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGUCCACCCAUGUAAGGUGGUUGUCCGGCCCCGUAGGGCGGAGCUUCUCCGUAC ...((.((((...((((.....))))..))))(((.((((...((((.((((.(((..((((....))))..))).))))..))))..)))))))......))((((((...)))))).. ( -51.00) >DroEre_CAF1 79079 108 + 1 CCACCGGGACCACCUCCAUAAGGUG------------GAGCACCCGCAUAAGGUGGCUGGCCACCUGGACCACCCACGUAAGGUGGUUGUCCGGCUCCGUAGGGCUGAGCUUCUCCGUAC ....(((((((((((......((((------------(.((....))...((((((....))))))...)))))......))))))))(..((((((....))))))..)....)))... ( -45.60) >DroYak_CAF1 78455 90 + 1 CCACCUGGACCA------------------------------CCCACAUAAGGUGGCUGUCCACCUGGACCACCCACGUAAGGUGGUUGUCCGGCGCCGUAGGGCGGCGCUUCUCCGUAC ......((((..------------------------------........((((((....)))))).(((((((.......)))))))))))(((((((.....)))))))......... ( -37.80) >consensus CCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAC ......((((...........(((((...(((....)))))))).......(((((....)))))..(((((((.......)))))))))))(((((((.....)))))))......... (-32.83 = -34.94 + 2.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:52 2006