
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,190,747 – 10,190,870 |
| Length | 123 |
| Max. P | 0.970758 |

| Location | 10,190,747 – 10,190,846 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190747 99 + 23771897 ACACAA---UGAUCGGGCUAAUUGUGCAAAAAUGU--------UUUUGCAAAUAUUUUGUUAACAAAUUUUGCGGCAUUUUAAGCCACAGAAAUGCAAUUAGC-CGUGAAC ......---...(((((((((((((((((((.(((--------(...((((.....)))).))))..))))))(((.......)))........)))))))))-).))).. ( -29.10) >DroVir_CAF1 95204 106 + 1 AUACAA---UUGUCAUGCUAAUUGUAUAAAAAUGA-UAAAGAGUUUUGCAAAUAUUUUGUUAACAAAUUGUGUGGCAAUUUAGGCCACAAAAAUGCAAUUAGC-UGUACAC ......---.(((((.(((((((((((.....(((-(((((.(((.....))).))))))))........((((((.......))))))...)))))))))))-)).))). ( -27.90) >DroPse_CAF1 76843 107 + 1 AUACAA---UGACCGAGCUAAUUGUGCAAAAAUGUGUGUGUUUUUCUGCAAAUAUUUUGUUAACAAAUUUUGCGGCCAUUUAAACCACAGAAAUGCAAUUAGC-CAUGAAC ......---.....(.((((((((((((....)))(((.((((..(((((((...((((....)))).)))))))......)))))))......)))))))))-)...... ( -24.80) >DroWil_CAF1 97216 99 + 1 GCACAA---UGACCAAACUAAUUGCGCAAAAAUGU--------U-UGGCAAAUAUUUUGUUAACAAAUUGUGUGGCAAUUUAAGCCACAGAAAUGCAAUUAGCCCAUAAGC ......---........(((((((((......(((--------(-.(((((.....)))))))))..((.((((((.......)))))).)).)))))))))......... ( -24.10) >DroAna_CAF1 65247 102 + 1 AUACAUUCCUGUGCUGGCUAGUUGUGCAAACAUGU--------UAUUGCAAAUAUUUUGUUAACAAAUUUUGUGGCCUUUUAAGCCACAGAAACGCAAUUAGC-CGUGAAC .((((....))))..(((((((((((......(((--------((..((((.....)))))))))..(((((((((.......))))))))).))))))))))-)...... ( -32.40) >DroPer_CAF1 77081 107 + 1 AUACAA---UGACCGAGCUAAUUGUGCAAAAAUGUGUGUGUUUUUCUGCAAAUAUUUUGUUAACAAAUUUUGCGGCCAUUUAAACCACAGAAAUGCAAUUAGC-CAUGAAC ......---.....(.((((((((((((....)))(((.((((..(((((((...((((....)))).)))))))......)))))))......)))))))))-)...... ( -24.80) >consensus AUACAA___UGACCGAGCUAAUUGUGCAAAAAUGU________UUUUGCAAAUAUUUUGUUAACAAAUUUUGCGGCAAUUUAAGCCACAGAAAUGCAAUUAGC_CAUGAAC ................((((((((((.....................(((((...((((....)))).)))))(((.......))).......))))))))))........ (-17.39 = -17.42 + 0.03)
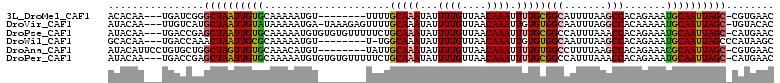

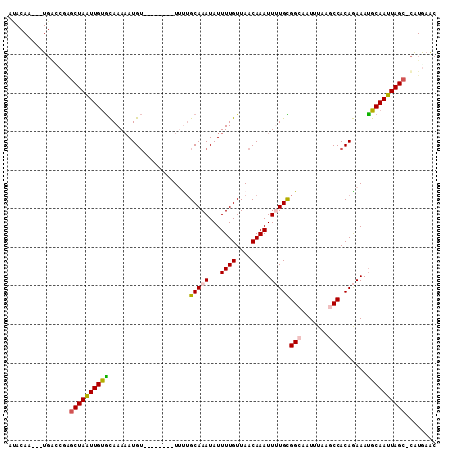
| Location | 10,190,747 – 10,190,846 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -19.35 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190747 99 - 23771897 GUUCACG-GCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAAAAUUUGUUAACAAAAUAUUUGCAAAA--------ACAUUUUUGCACAAUUAGCCCGAUCA---UUGUGU ......(-((((((((......((((((.......))))))...((((....)))).....(((((((--------....))))))))))))))))......---...... ( -27.30) >DroVir_CAF1 95204 106 - 1 GUGUACA-GCUAAUUGCAUUUUUGUGGCCUAAAUUGCCACACAAUUUGUUAACAAAAUAUUUGCAAAACUCUUUA-UCAUUUUUAUACAAUUAGCAUGACAA---UUGUAU .(((.((-((((((((......((((((.......))))))...(((((.............)))))........-...........)))))))).))))).---...... ( -21.12) >DroPse_CAF1 76843 107 - 1 GUUCAUG-GCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAAAAUUUGUUAACAAAAUAUUUGCAGAAAAACACACACAUUUUUGCACAAUUAGCUCGGUCA---UUGUAU ......(-((((((((......(((((((.....)))))))...((((....)))).....((((((((..........)))))))))))))))))......---...... ( -23.60) >DroWil_CAF1 97216 99 - 1 GCUUAUGGGCUAAUUGCAUUUCUGUGGCUUAAAUUGCCACACAAUUUGUUAACAAAAUAUUUGCCA-A--------ACAUUUUUGCGCAAUUAGUUUGGUCA---UUGUGC .....(..(((((((((.....((((((.......))))))(((..((((..(((.....)))...-)--------)))...))).)))))))))..)....---...... ( -24.70) >DroAna_CAF1 65247 102 - 1 GUUCACG-GCUAAUUGCGUUUCUGUGGCUUAAAAGGCCACAAAAUUUGUUAACAAAAUAUUUGCAAUA--------ACAUGUUUGCACAACUAGCCAGCACAGGAAUGUAU ......(-((((.(((.((...(((((((.....)))))))((((.(((((.(((.....)))...))--------))).)))))).))).)))))...(((....))).. ( -25.20) >DroPer_CAF1 77081 107 - 1 GUUCAUG-GCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAAAAUUUGUUAACAAAAUAUUUGCAGAAAAACACACACAUUUUUGCACAAUUAGCUCGGUCA---UUGUAU ......(-((((((((......(((((((.....)))))))...((((....)))).....((((((((..........)))))))))))))))))......---...... ( -23.60) >consensus GUUCACG_GCUAAUUGCAUUUCUGUGGCUUAAAUGGCCACAAAAUUUGUUAACAAAAUAUUUGCAAAA________ACAUUUUUGCACAAUUAGCUCGGUCA___UUGUAU .....((.((((((((......((((((.......))))))...((((....)))).....(((((((............))))))))))))))).))............. (-19.35 = -18.88 + -0.47)


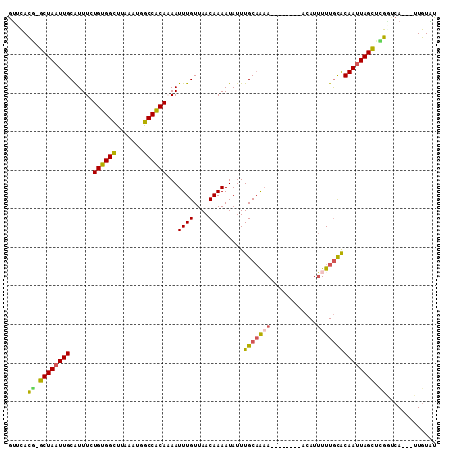
| Location | 10,190,777 – 10,190,870 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -17.48 |
| Consensus MFE | -14.00 |
| Energy contribution | -13.37 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190777 93 - 23771897 -ACU-AAAAUUUUG-UCGGUUAAUUACGUUCACGGCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAAAAUUUGUUAACAAAAUAUUUGCAAAA--------AC -...-........(-(((..............))))...(((((....((((((.......))))))...((((....)))).....)))))..--------.. ( -17.94) >DroVir_CAF1 95234 99 - 1 -AUA-CUCAUAU--UUCAGUUAAUUGCGUGUACAGCUAAUUGCAUUUUUGUGGCCUAAAUUGCCACACAAUUUGUUAACAAAAUAUUUGCAAAACUCUUUA-UC -...-...((((--((..((((((....(((...((.....))......(((((.......))))))))....)))))).))))))...............-.. ( -16.60) >DroPse_CAF1 76873 104 - 1 CAUUCCUCAUUUUGUUCGGUUAAUUACGUUCAUGGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAAAAUUUGUUAACAAAAUAUUUGCAGAAAAACACACAC ..(((..((...((((..((((((....((..((((((((..((....))..)).....))))))..))....))))))..))))..))..))).......... ( -17.20) >DroEre_CAF1 71449 93 - 1 -ACU-CACAUUUUG-UCGGUUAAUUACAUUCACGGCUAAUUGCAUUUUUGUGGCUUCAAAUGCCGCAAAAUUUGUUAACAUAAUAUUUGCAAAA--------AC -...-........(-(((..............))))...(((((.(((((((((.......)))))))))..((....)).......)))))..--------.. ( -18.44) >DroAna_CAF1 65280 95 - 1 CAUU-CUCAUUUUAUUUGGUUAAUUACGUUCACGGCUAAUUGCGUUUCUGUGGCUUAAAAGGCCACAAAAUUUGUUAACAAAAUAUUUGCAAUA--------AC ....-.......(((((.((((((..((....))((.....)).....(((((((.....)))))))......)))))).))))).........--------.. ( -17.50) >DroPer_CAF1 77111 104 - 1 CAUUCCUCAUUUUGUUCGGUUAAUUACGUUCAUGGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAAAAUUUGUUAACAAAAUAUUUGCAGAAAAACACACAC ..(((..((...((((..((((((....((..((((((((..((....))..)).....))))))..))....))))))..))))..))..))).......... ( -17.20) >consensus _AUU_CUCAUUUUGUUCGGUUAAUUACGUUCACGGCUAAUUGCAUUUCUGUGGCUUAAAAGGCCGCAAAAUUUGUUAACAAAAUAUUUGCAAAA________AC .......................................(((((....((((((.......))))))...((((....)))).....)))))............ (-14.00 = -13.37 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:47 2006