
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,190,435 – 10,190,595 |
| Length | 160 |
| Max. P | 0.915468 |

| Location | 10,190,435 – 10,190,555 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -30.28 |
| Energy contribution | -32.20 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190435 120 + 23771897 UUGAAGAACUUUGAUACCAUGGAACUUCCAUCUAUAUCGGAGCUAUCGUUGCUGCUGGACAAGAGCACCCAGGUGACCAAGCGCAGGAGCUCAAUUGAAAAGGAGCGAAGAUCCUUCAAG ((((((..((((((((..((((.....))))...))))))))..(((.(((((.((...(((((((.((...(((......))).)).))))..)))...)).))))).))).)))))). ( -36.40) >DroSec_CAF1 66891 120 + 1 UCGAAGAACUUUGAUACCAUGGUACUUCCAUCUAUAUCGGAGCUAUCGUUGCUGCUGGACAAGAGCUCCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAG ..((((..((((((((..((((.....))))...))))))))..(((.(((((.((.((((.(((((((...(((......))).)))))))...)))).)).))))).))).))))... ( -42.40) >DroSim_CAF1 70851 120 + 1 UUGAAGAACUUUGAUACCAUGUUACUUCCAUCUAUAUCGGAGCUAUCGUUGCUGCUGGACAAGAGCUCCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAG ((((((..((((((((..(((.......)))...))))))))..(((.(((((.((.((((.(((((((...(((......))).)))))))...)))).)).))))).))).)))))). ( -39.60) >DroEre_CAF1 71108 120 + 1 GUGAGGAACUUCGAUAACCUGGUUCUGCCAUCCAUAUCGGAGCUAUCGCUGCUGCUGGAGAAGAGCGUCCAGGUGAACAAGCGCAGGAACUCAAGUAUCAAGGAGCGAAGAUCCUUCAAG .((((((.(((((....(((.(((((....((((...((((((....))).))).))))..))))).(((..(((......))).)))............)))..))))).))).))).. ( -34.40) >DroYak_CAF1 70830 120 + 1 UUGAAGAACUUCAACAACCUGGUACUUCCAUCCAUGUCGGAGCUAUCGUUGCUGCUGGAGAAGAGCACCCAACUGACCAGGCGCCGGAGCUCAAGUGCCAAGGAGCGAAGAUCCUUCAAG ((((((..((((.....((((((((((...((((.((.(.(((....))).).))))))...((((.((...((....)).....)).))))))))))).)))...))))...)))))). ( -35.70) >consensus UUGAAGAACUUUGAUACCAUGGUACUUCCAUCUAUAUCGGAGCUAUCGUUGCUGCUGGACAAGAGCACCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAG ((((((..((((((((..((((.....))))...))))))))..(((.(((((.((.((((.((((.((...(((......))).)).))))...)))).)).))))).))).)))))). (-30.28 = -32.20 + 1.92)

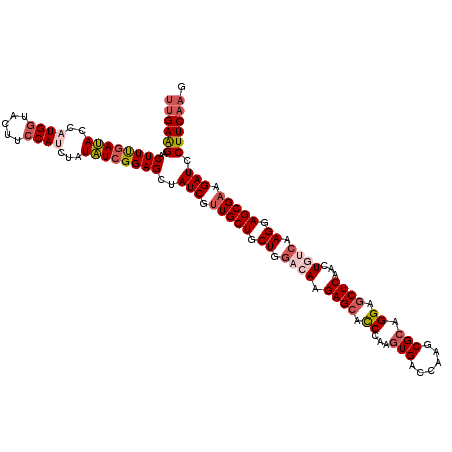
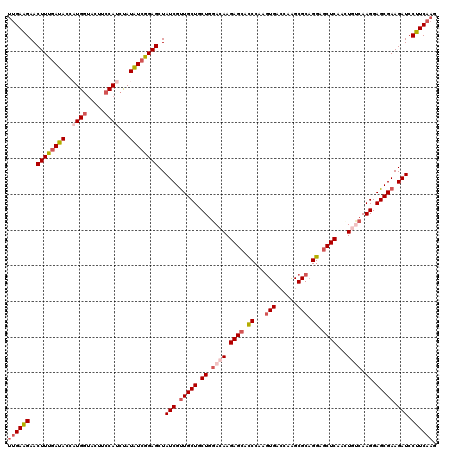
| Location | 10,190,475 – 10,190,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -32.10 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190475 120 + 23771897 AGCUAUCGUUGCUGCUGGACAAGAGCACCCAGGUGACCAAGCGCAGGAGCUCAAUUGAAAAGGAGCGAAGAUCCUUCAAGUUCGCCUGGGCUGCUCCUUUGGCGAAGCAGGAAAAAUACA .(.(((..((.(((((...(((((((((((((((((.(.(((......)))...((((..((((.......))))))))).))))))))).)))))..)))....))))).))..)))). ( -44.10) >DroSec_CAF1 66931 120 + 1 AGCUAUCGUUGCUGCUGGACAAGAGCUCCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAGUUCGCCUGGGUUGCUCCUUUGGCGAAGCAGGAAAAAUACA .....((.(((((.((.((((.(((((((...(((......))).)))))))...)))).)).))))).))((((.(...((((((.(((.....)))..))))))).))))........ ( -43.30) >DroSim_CAF1 70891 120 + 1 AGCUAUCGUUGCUGCUGGACAAGAGCUCCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAGUUCGCCUGGGCUGCUCCUUUGGCGAAGCAGGAAAAAUACA .(.(((..((.(((((......(((((((...(((......))).)))))))...(((((((((((((((...)))).(((((....)))))))))).)))))).))))).))..)))). ( -43.50) >DroEre_CAF1 71148 120 + 1 AGCUAUCGCUGCUGCUGGAGAAGAGCGUCCAGGUGAACAAGCGCAGGAACUCAAGUAUCAAGGAGCGAAGAUCCUUCAAGUUCGCCUGGGCUGCUCCUUUAACGAAGCAGGAGAAAUACU ...(((..((.(((((..(((.((((((((((((((((..((.(..((((....)).))...).))((((...))))..)))))))))))).)))).))).....))))).))..))).. ( -45.70) >DroYak_CAF1 70870 120 + 1 AGCUAUCGUUGCUGCUGGAGAAGAGCACCCAACUGACCAGGCGCCGGAGCUCAAGUGCCAAGGAGCGAAGAUCCUUCAAGUUCGCCCUGGCUGCUCCUCUGCCAAAGCAGAAAAAAUACU ........(((((..((((((.(((((.((.........(((((..(....)..))))).(((.(((((((....))...)))))))))).))))).))).))).))))).......... ( -40.80) >consensus AGCUAUCGUUGCUGCUGGACAAGAGCACCCAAGUGACCAAGCGCAGGAGCUCAACUGUCAAGGAGCGAAGAUCCUUCAAGUUCGCCUGGGCUGCUCCUUUGGCGAAGCAGGAAAAAUACA ...(((..((.(((((......((((.((...(((......))).)).))))...(((((((((((((((...))))......((....)).)))))).))))).))))).))..))).. (-32.10 = -32.62 + 0.52)



| Location | 10,190,475 – 10,190,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10190475 120 - 23771897 UGUAUUUUUCCUGCUUCGCCAAAGGAGCAGCCCAGGCGAACUUGAAGGAUCUUCGCUCCUUUUCAAUUGAGCUCCUGCGCUUGGUCACCUGGGUGCUCUUGUCCAGCAGCAACGAUAGCU ..((((.((.(((((..((...(((((((.((((((.((....((((((.......))))))....(..(((......)))..))).)))))))))))))))..))))).)).))))... ( -40.90) >DroSec_CAF1 66931 120 - 1 UGUAUUUUUCCUGCUUCGCCAAAGGAGCAACCCAGGCGAACUUGAAGGAUCUUCGCUCCUUGACAGUUGAGCUCCUGCGCUUGGUCACUUGGGAGCUCUUGUCCAGCAGCAACGAUAGCU ..((((.((.((((((((((...((......)).))))).......((((....((((((((((....((((......)))).))))...))))))....))))))))).)).))))... ( -37.70) >DroSim_CAF1 70891 120 - 1 UGUAUUUUUCCUGCUUCGCCAAAGGAGCAGCCCAGGCGAACUUGAAGGAUCUUCGCUCCUUGACAGUUGAGCUCCUGCGCUUGGUCACUUGGGAGCUCUUGUCCAGCAGCAACGAUAGCU ..((((.((.(((((......(((((((.((....))......((((...)))))))))))(((((..(((((((((.(........).)))))))))))))).))))).)).))))... ( -40.10) >DroEre_CAF1 71148 120 - 1 AGUAUUUCUCCUGCUUCGUUAAAGGAGCAGCCCAGGCGAACUUGAAGGAUCUUCGCUCCUUGAUACUUGAGUUCCUGCGCUUGUUCACCUGGACGCUCUUCUCCAGCAGCAGCGAUAGCU ........(((((((..(((..(((((.((((((((.((((..(.((((.....((((..........))))))))...)..)))).)))))..))))))))..)))))))).))..... ( -37.60) >DroYak_CAF1 70870 120 - 1 AGUAUUUUUUCUGCUUUGGCAGAGGAGCAGCCAGGGCGAACUUGAAGGAUCUUCGCUCCUUGGCACUUGAGCUCCGGCGCCUGGUCAGUUGGGUGCUCUUCUCCAGCAGCAACGAUAGCU ..((((.((.(((((..((.(((((((((.((((((((((..(.....)..)))))))((.(((.((.(....).)).))).)).....))).)))))))))))))))).)).))))... ( -44.00) >consensus UGUAUUUUUCCUGCUUCGCCAAAGGAGCAGCCCAGGCGAACUUGAAGGAUCUUCGCUCCUUGACACUUGAGCUCCUGCGCUUGGUCACUUGGGAGCUCUUGUCCAGCAGCAACGAUAGCU ..((((.((.(((((..((...((((((.(((((((.((.(..(.((((.....((((..........))))))))...)..).)).)))))))))))))))..))))).)).))))... (-31.86 = -32.06 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:44 2006