
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,003,680 – 1,003,780 |
| Length | 100 |
| Max. P | 0.868894 |

| Location | 1,003,680 – 1,003,780 |
|---|---|
| Length | 100 |
| Sequences | 6 |
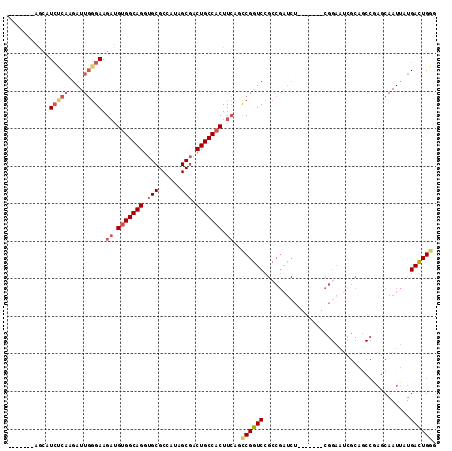
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -22.57 |
| Energy contribution | -23.51 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
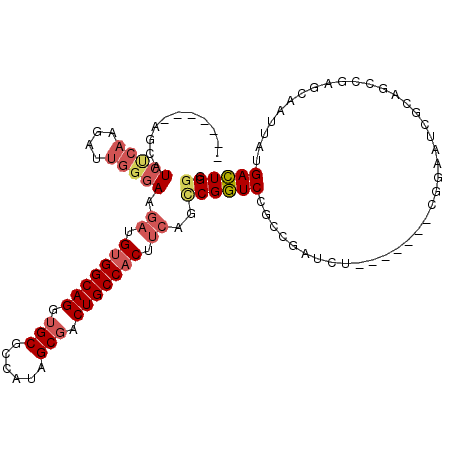
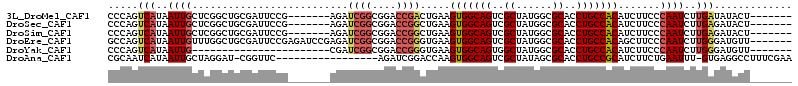
>3L_DroMel_CAF1 1003680 100 + 23771897 -------AGUAUAUCAAGAUUGGGAAGAUGUGGCAGGUGCGCCAUAGCGACUGCCACUUCAGUCGGUCCGCCGAUCU-------CGGAAUCGCAGCCGAGCAAUUAUGACUGGG -------......(((.(((((....((.(((((((...(((....))).))))))).)).(((((....)))))((-------(((........)))))))))).)))..... ( -36.50) >DroSec_CAF1 21888 100 + 1 -------AGUAUCUCAAGAUUGGGAAGAUGUGGCAGGUGCGCCAUAGCGACUGCCACUUCAGCCGGUCCGCCGAUCU-------CGGAAUCGCAGCCGAGCAAUUAUGACUGGG -------....(((((....))))).((.(((((((...(((....))).))))))).))..((((((.......((-------(((........))))).......)))))). ( -38.44) >DroSim_CAF1 22091 100 + 1 -------AGUAUCUCAAGAUUGGGAAGAUGUGGCAGGUGCGCCAUAGCGACUGCCACUUCAGCCGGUCCGCCGAUCU-------CGGAAUCGCAGCCGAGCAAUUAUGACUGGG -------....(((((....))))).((.(((((((...(((....))).))))))).))..((((((.......((-------(((........))))).......)))))). ( -38.44) >DroEre_CAF1 23547 107 + 1 -------AACAUCCCAAGAUUGGGAAGCUGUGGCAGGUGCGCCAUAGCGACUGCCACUUCACCCGGUCCGCCGAUCUCGGAUCUCGGAAUCGCAGCCAAACAAUUAUGACUGGC -------....(((((....))))).((.(((((((...(((....))).)))))))....((.((((((.......))))))..))....)).((((..((....))..)))) ( -38.10) >DroYak_CAF1 22550 84 + 1 -------AACAUCCCAAGAUUGGGAAGAUGUGGCAGGUGCGCCAUAGCCACUGCCACUUCACCCGGUCCGCCGAUCG-----------------------CAAUUAUGACUGGG -------....(((((....))))).((.(((((((..((......))..))))))).)).(((((((.((.....)-----------------------)......))))))) ( -33.80) >DroAna_CAF1 27305 95 + 1 UUCGAAAGGCCUCAC-AAAUUCAGAAGAUGCGGCAGGUGCGCUAUAGCGACUGCCACUUGGUCCGAUCU-----------------GAACCG-AUCCUAGCAAUUAUGAUUGCG .((....))......-...((((((.((((.(((((...(((....))).))))).)...)))...)))-----------------)))...-......(((((....))))). ( -25.20) >consensus _______AGCAUCUCAAGAUUGGGAAGAUGUGGCAGGUGCGCCAUAGCGACUGCCACUUCAGCCGGUCCGCCGAUCU_______CGGAAUCGCAGCCGAGCAAUUAUGACUGGG ...........(((((....))))).((.(((((((.(((......))).))))))).))..((((((.......................................)))))). (-22.57 = -23.51 + 0.95)

| Location | 1,003,680 – 1,003,780 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
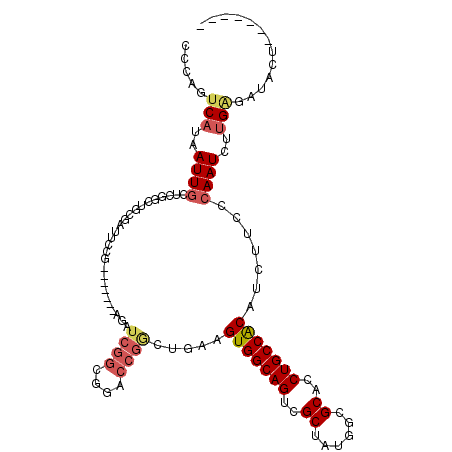
>3L_DroMel_CAF1 1003680 100 - 23771897 CCCAGUCAUAAUUGCUCGGCUGCGAUUCCG-------AGAUCGGCGGACCGACUGAAGUGGCAGUCGCUAUGGCGCACCUGCCACAUCUUCCCAAUCUUGAUAUACU------- ....((((..((((.((((((((((((...-------.)))).)))).))))..((.(((((((.(((....)))...))))))).))....))))..)))).....------- ( -35.10) >DroSec_CAF1 21888 100 - 1 CCCAGUCAUAAUUGCUCGGCUGCGAUUCCG-------AGAUCGGCGGACCGGCUGAAGUGGCAGUCGCUAUGGCGCACCUGCCACAUCUUCCCAAUCUUGAGAUACU------- .....(((..((((...((((((((((...-------.)))).)))).))((..((.(((((((.(((....)))...))))))).))..))))))..)))......------- ( -33.70) >DroSim_CAF1 22091 100 - 1 CCCAGUCAUAAUUGCUCGGCUGCGAUUCCG-------AGAUCGGCGGACCGGCUGAAGUGGCAGUCGCUAUGGCGCACCUGCCACAUCUUCCCAAUCUUGAGAUACU------- .....(((..((((...((((((((((...-------.)))).)))).))((..((.(((((((.(((....)))...))))))).))..))))))..)))......------- ( -33.70) >DroEre_CAF1 23547 107 - 1 GCCAGUCAUAAUUGUUUGGCUGCGAUUCCGAGAUCCGAGAUCGGCGGACCGGGUGAAGUGGCAGUCGCUAUGGCGCACCUGCCACAGCUUCCCAAUCUUGGGAUGUU------- (((((.((....)).)))))....(((((((((((((.......))))..(((.(..(((((((.(((....)))...)))))))..)..)))..)))))))))...------- ( -43.20) >DroYak_CAF1 22550 84 - 1 CCCAGUCAUAAUUG-----------------------CGAUCGGCGGACCGGGUGAAGUGGCAGUGGCUAUGGCGCACCUGCCACAUCUUCCCAAUCUUGGGAUGUU------- (((((......(((-----------------------(.....))))...(((.((.(((((((((((....)).)).))))))).))..)))....))))).....------- ( -31.40) >DroAna_CAF1 27305 95 - 1 CGCAAUCAUAAUUGCUAGGAU-CGGUUC-----------------AGAUCGGACCAAGUGGCAGUCGCUAUAGCGCACCUGCCGCAUCUUCUGAAUUU-GUGAGGCCUUUCGAA .((..((((((...(.(((((-.(((((-----------------.....)))))..(((((((.(((....)))...))))))))))))..)...))-)))).))........ ( -30.40) >consensus CCCAGUCAUAAUUGCUCGGCUGCGAUUCCG_______AGAUCGGCGGACCGGCUGAAGUGGCAGUCGCUAUGGCGCACCUGCCACAUCUUCCCAAUCUUGAGAUACU_______ .....(((..((((..........................((((....)))).....(((((((..((......))..))))))).......))))..)))............. (-18.76 = -19.52 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:43 2006