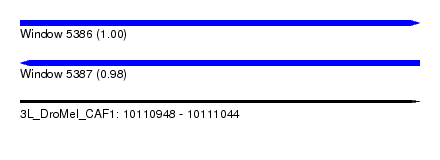
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,110,948 – 10,111,044 |
| Length | 96 |
| Max. P | 0.999632 |

| Location | 10,110,948 – 10,111,044 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -24.58 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10110948 96 + 23771897 UGUUUCCAUGUUGGUAACUGUGUGCAGUGCGUGUUCUAUAUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACACAAACAGCAUACACAG .......((((((.....((((((.(((((.((((.((((.((((((....)))))))))).)))).).))))...))))))..))))))...... ( -24.80) >DroSec_CAF1 49916 94 + 1 UGUAUCCCUGUUGGUAACUGUGUGCAGUGCGUGUUGUAUAUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACACAAACAGCAUACAC-- (((((..(((((......((((((.(((((.((((.((((.((((((....)))))))))).)))).).))))...))))))))))).))))).-- ( -24.50) >DroSim_CAF1 50953 94 + 1 UGUAUCCCUGUUGGUAACUGUGUGCAGUGCGUGUUGUAUAUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACACAAACAGCAUACAC-- (((((..(((((......((((((.(((((.((((.((((.((((((....)))))))))).)))).).))))...))))))))))).))))).-- ( -24.50) >DroEre_CAF1 54620 94 + 1 UGUAUCCCUCUUGGCAACUGUGUGCAGUGCGCGUUGUAUGUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACACAAACAGCGCACAC-- (((..........(((.(((....))))))(((((((.(((((((((....((((.(((.....)))))))..))))))))).)))))))))).-- ( -35.20) >DroYak_CAF1 52797 94 + 1 UGUAUCCCUCUCGGUAGCUGUGUGCAGUGCGUGUUGUGUAUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACAGAGACAGCAUGCAC-- ..((((......))))((.....)).(((((((((((.(.(((((((....((((.(((.....)))))))..))))))).).)))))))))))-- ( -31.60) >consensus UGUAUCCCUGUUGGUAACUGUGUGCAGUGCGUGUUGUAUAUGUGUUAUCAUUGGCAUUGUAAAACAAGUCACUUAACACACAAACAGCAUACAC__ (((..........(((.(((....))))))(((((((.(.(((((((....((((.(((.....)))))))..))))))).).))))))))))... (-24.58 = -23.98 + -0.60)



| Location | 10,110,948 – 10,111,044 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10110948 96 - 23771897 CUGUGUAUGCUGUUUGUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACAUAUAGAACACGCACUGCACACAGUUACCAACAUGGAAACA .((((((((((((((((((((((..((.((((.....))).).))....))))))))...))))).))).))))))..(((.((.....)).))). ( -25.00) >DroSec_CAF1 49916 94 - 1 --GUGUAUGCUGUUUGUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACAUAUACAACACGCACUGCACACAGUUACCAACAGGGAUACA --(((((((((((((((((((((..((.((((.....))).).))....)))))))))...)))).))).)))))...((..((....))...)). ( -22.90) >DroSim_CAF1 50953 94 - 1 --GUGUAUGCUGUUUGUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACAUAUACAACACGCACUGCACACAGUUACCAACAGGGAUACA --(((((((((((((((((((((..((.((((.....))).).))....)))))))))...)))).))).)))))...((..((....))...)). ( -22.90) >DroEre_CAF1 54620 94 - 1 --GUGUGCGCUGUUUGUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACACAUACAACGCGCACUGCACACAGUUGCCAAGAGGGAUACA --(((((((.(((.(((((((((..((.((((.....))).).))....))))))))).))).)))))))........((..((.....))..)). ( -29.50) >DroYak_CAF1 52797 94 - 1 --GUGCAUGCUGUCUCUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACAUACACAACACGCACUGCACACAGCUACCGAGAGGGAUACA --(((((((((((...(((((...(((..(((((.............))))))))..)))))))).))).))))).......((....))...... ( -22.32) >consensus __GUGUAUGCUGUUUGUGUGUUAAGUGACUUGUUUUACAAUGCCAAUGAUAACACAUAUACAACACGCACUGCACACAGUUACCAACAGGGAUACA ..(((((((((((((((((((((..((.((((.....))).).))....)))))))))...)))).))).))))).......((.....))..... (-20.64 = -20.64 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:24 2006