| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,110,275 – 10,110,373 |
| Length | 98 |
| Max. P | 0.625340 |

| Location | 10,110,275 – 10,110,373 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -16.28 |
| Energy contribution | -17.62 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
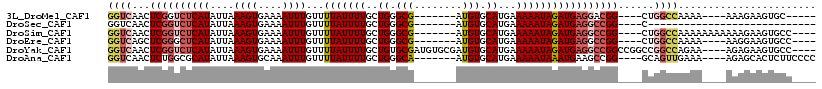
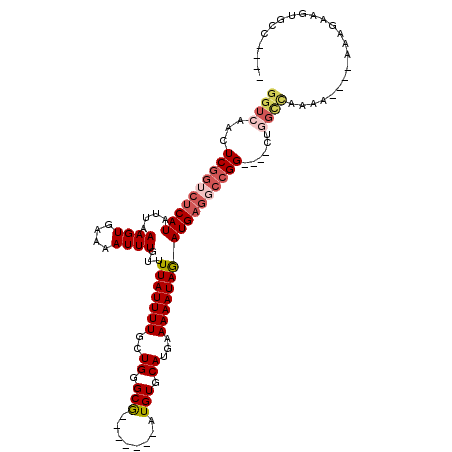
>3L_DroMel_CAF1 10110275 98 - 23771897 GGUCAACUCGGUCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGGGCG-------AUGUGCAUGAAAAAUAGAUGAGGACGG----CUGGCCAAAA----AAAGAAGUGC----- (((((.(.((..(((((....((((....))))...(((((((.....((.-------....))....))))))))))))..)))----.)))))....----..........----- ( -21.50) >DroSec_CAF1 49263 79 - 1 GGUCAACUCGGUCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGGGCG-------AUGUGCAUGAAAAAUAGAUGAGGCCGG----C---------------------------- .......((((((((((....((((....))))...(((((((.....((.-------....))....)))))))))))))))))----.---------------------------- ( -19.40) >DroSim_CAF1 50262 103 - 1 GGUCAACUCGGUCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGGGCG-------AUGUGCAUGAAAAAUAGAUGAGGCCGG----CUGGCCAAAAAAAAAAAGAAGUGCC---- (((((.(.(((((((((....((((....))))...(((((((.....((.-------....))....)))))))))))))))))----.)))))...................---- ( -27.30) >DroEre_CAF1 53968 99 - 1 GGUCAGCUCGGGCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGGGCG-------AUGUGCAUGAAAAAUAGAUGAGGCCGG----CUGGCCAAAA----AAGGAAGUGCC---- (((((((.(((.(((((....((((....))))...(((((((.....((.-------....))....)))))))))))).))))----))))))....----...........---- ( -32.00) >DroYak_CAF1 52089 110 - 1 GGUCAACUCGGUCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGUGCGAUGUGCGAUGUGCAUGAAAAAUAGAUGAGGCCGGCCGGCCGGCCAGAA----AGAGAAGUGCC---- ((((..(((((((((((..((((((((((.....))))))))))((((.....((((.....)))).....))))))))))))((((....))))....----.)))..).)))---- ( -35.40) >DroAna_CAF1 47489 103 - 1 GGUCAACUCUGGCGCAUAUUAAAGUGCAAAUUUGUUUUAUUUUGCUGGGCA-------AUGUGCAUGAAAAAUAAAUGAAGCCGG----GCAGUUGAAA----AGAGCACUCUUCCCC (((((((((((((((((......))))..(((((((((.((.(((......-------....))).)))))))))))...)))))----..)))))).(----(((....)))).)). ( -25.90) >consensus GGUCAACUCGGUCUCAUAUUAAAGUGAAAAUUUGUUUUAUUUUGCUGGGCG_______AUGUGCAUGAAAAAUAGAUGAGGCCGG____CUGGCCAAAA____AAAGAAGUGCC____ ((((...((((((((((....((((....))))...(((((((..((.(((........))).))...)))))))))))))))))......))))....................... (-16.28 = -17.62 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:22 2006