
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,110,045 – 10,110,240 |
| Length | 195 |
| Max. P | 0.991172 |

| Location | 10,110,045 – 10,110,136 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
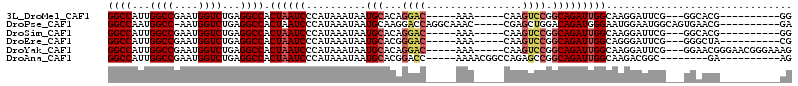
>3L_DroMel_CAF1 10110045 91 - 23771897 GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAA-----CAAGUCCGGCAGAUUGGCAAGGAUUCG---GGCACG----------GG ((((...)))).......(((..(((.(.(((((.........(((...((((-----...-----...)))).))).........))))).)---))).))----------). ( -28.87) >DroPse_CAF1 53723 98 - 1 GGCCAAUGGCC-AAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCAAGGACCAGGCAAAC-----CGAGCUGGACAGAUGGGAAUGGAAUGGCAGUGAACG----------GA .((((.(((((-..........)))))(((.((((((.......((..((..........)-----)..)).......)))))).)))..))))........----------.. ( -28.34) >DroSim_CAF1 50036 91 - 1 GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAA-----CAAGUCCGGCAGAUUGGCAAGGAUUCG---GGCACG----------GG ((((...)))).......(((..(((.(.(((((.........(((...((((-----...-----...)))).))).........))))).)---))).))----------). ( -28.87) >DroEre_CAF1 53723 91 - 1 GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACGGGAC-----AAA-----CAAGUCCGGCAGAUUGGCAGGGAUUCG---GGGCUA----------CG (((((((.....))))))).(.((((.((((((((........(((...((((-----...-----...)))).)))........)))))).)---))))).----------). ( -32.19) >DroYak_CAF1 51846 101 - 1 GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAA-----CAAGUCCGGCAGAUUGGCAAGGAUUCG---GGAACGGGAACGGGAAAG ((((...((((....))))...)))).....((((........(((...((((-----...-----...)))).))).........(..((((---....))))..)))))... ( -34.30) >DroAna_CAF1 47269 91 - 1 GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACGGACC-----AAAACGGCCAGAGCCGGCAGAUUGGCAAGACGGC--------GA----------AG .(((.(((((((..(((((((..((...................)).))))))-----)...))))))).(((((....))))).....)))--------..----------.. ( -32.91) >consensus GGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC_____AAA_____CAAGUCCGGCAGAUUGGCAAGGAUUCG___GGAACG__________GG ((((...((((....))))...)))).((((((..........(((...((((................)))).)))))))))............................... (-17.17 = -17.72 + 0.56)


| Location | 10,110,072 – 10,110,169 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10110072 97 + 23771897 CGGACUUGUUU-----GUCCUGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGGCCACCCAAGCCAAAGCCCAGG-------CC .((.((((..(-----(.((((.((((....((......))((((((...(.((....)).)...)))))))))))))).))..))))))...((....)-------). ( -33.20) >DroPse_CAF1 53753 96 + 1 CCAGCUCGGUUUGCCUGGUCCUUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUU-GGCCAUUGGCCAUUGGCCGUC--C-----GGCAACGCUGCGCUG-----CC .((((.(((((((((.........(((.....)))((((.(((((((..........-)))))))((((...)))))))--)-----))))).)))).))))-----.. ( -37.70) >DroSec_CAF1 49055 102 + 1 CGGACUUGUUU-----GUCCUGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAAGCCCAGGACCCAAGCC .((.((((...-----((((((.((........(((.((..((((((...(.((....)).)...)))))))).)))((--.......))...)).)))))).)))))) ( -35.40) >DroSim_CAF1 50063 83 + 1 CGGACUUGUUU-----GUCCUGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAG-------------------CC .((.((((...-----((((((.((((....((......))((((((...(.((....)).)...))))))))))))))--)).))))-------------------)) ( -30.60) >DroEre_CAF1 53750 101 + 1 CGGACUUGUUU-----GUCCCGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAAGCCCAAG-CCCAAGCC .((.((((...-----((((((((.......))))))))..((((((...(.((....)).)...))))))(.((..(.--...)..)))......))))-.))..... ( -28.90) >DroYak_CAF1 51883 89 + 1 CGGACUUGUUU-----GUCCUGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAGGCC------------- .((.((((...-----((((((.((((....((......))((((((...(.((....)).)...))))))))))))))--)).))))))......------------- ( -32.30) >consensus CGGACUUGUUU_____GUCCUGUGCAUUAUUUAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG__ACCCAAGCCAAAGCCCAGG_______CC ....((((........((((((((.......))))))))..((((((...(.((....)).)...))))))....)))).............................. (-19.26 = -19.15 + -0.11)


| Location | 10,110,072 – 10,110,169 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -21.90 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
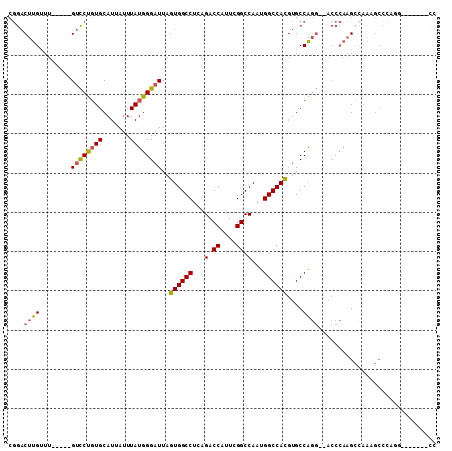
>3L_DroMel_CAF1 10110072 97 - 23771897 GG-------CCUGGGCUUUGGCUUGGGUGGCCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAACAAGUCCG ((-------((((((((((((((..(((((((........)))))))))))))..))))).))))).......................((((-----......)))). ( -39.40) >DroPse_CAF1 53753 96 - 1 GG-----CAGCGCAGCGUUGCC-----G--GACGGCCAAUGGCCAAUGGCC-AAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCAAGGACCAGGCAAACCGAGCUGG ((-----(((((...)))))))-----(--(...(((..(((((...))))-).((((((...((...................))..)))))))))...))....... ( -34.61) >DroSec_CAF1 49055 102 - 1 GGCUUGGGUCCUGGGCUUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAACAAGUCCG ((((((.((((((.((.(((...((((.--..((((....(((((((.....)))))))....)))).....))))....))).)).))))))-----...)))))).. ( -43.20) >DroSim_CAF1 50063 83 - 1 GG-------------------CUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAACAAGUCCG ((-------------------((((.((--(((((((.((((((...((((....))))...))))))((......)).....))).))))))-----...)))))).. ( -36.10) >DroEre_CAF1 53750 101 - 1 GGCUUGGG-CUUGGGCUUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACGGGAC-----AAACAAGUCCG .(((.(((-((..(((....)))..)))--)).)))..((((((...((((....))))...)))))).....................((((-----......)))). ( -43.10) >DroYak_CAF1 51883 89 - 1 -------------GGCCUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC-----AAACAAGUCCG -------------......((((((.((--(((((((.((((((...((((....))))...))))))((......)).....))).))))))-----...)))))).. ( -36.10) >consensus GG_______CCUGGGCUUUGGCUUGGGU__CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUAAAUAAUGCACAGGAC_____AAACAAGUCCG ............((((.(((..........(((((((.((((((...((((....))))...))))))((......)).....))).))))..........))))))). (-21.90 = -23.10 + 1.20)

| Location | 10,110,098 – 10,110,207 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -35.21 |
| Consensus MFE | -24.13 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
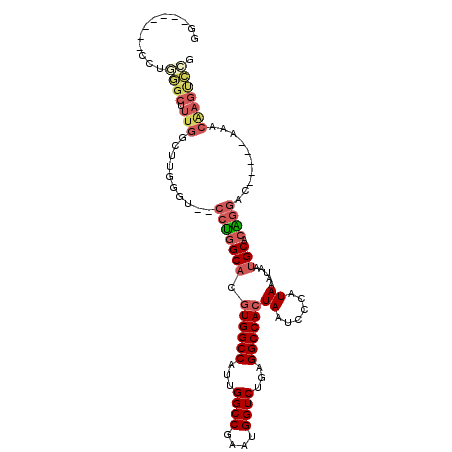
>3L_DroMel_CAF1 10110098 109 + 23771897 UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGGCCACCCAAGCCAAAGCCCAGG-------CCC-AAG-CCCAAGCCCAAAGAGGCUCUGGCCUCAAAACUGA ..((((.....((((....((....))((((..((((.....))))))))......))))...))))((-------((.-.((-(((.........).))))..)))).......... ( -37.50) >DroPse_CAF1 53784 104 + 1 UAUGGGAUUAGUGGCCUCAGACCAUU-GGCCAUUGGCCAUUGGCCGUC--C-----GGCAACGCUGCGCUG-----CCA-CGUGCCACGGCCGAACCUUAUUUUGGCCUCAAAACUGA ....((.(((((((((..........-)))))))))))..(((((((.--.-----((((.((...)).))-----)))-)).)))).(((((((......))))))).......... ( -38.90) >DroSec_CAF1 49081 114 + 1 UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAAGCCCAGGACCCAAGCCA-AAG-CCCAGGCCCAAACAGGCUCUGGCCUCAAAACUGA ..((((.....(((((...........))))).((((.....))))..--.)))).((((.((((..((.((...((..-..)-)...)).)).....)))).))))........... ( -34.50) >DroSim_CAF1 50089 95 + 1 UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAG-------------------CCA-AGG-CCCAGGCCCAAACAGGCUCUGGCCUCAAAACUGA ..((((....((((((...(.((....)).)...))))))..(((((.--.((....-------------------...-.((-(....)))......))..)))))))))....... ( -33.44) >DroEre_CAF1 53776 114 + 1 UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAAGCCCAAG-CCCAAGCCCAAGC-UCCAGGCCCAAACUGGCUCUGGCCUCAAAACUGA ..((((.....(((((...........))))).((((.....))))..--.)))).((((.((((...(-((..(((....))-)...))).......)))).))))........... ( -35.30) >DroYak_CAF1 51909 102 + 1 UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG--ACCCAAGCCAAGGCC-------------CAAAC-UCCGUGCCCAAACUGGCUCUGGCCUCAAAACUGA ..((((....((((((...(.((....)).)...))))))..(((((.--.((..((....(((.-------------(....-...).)))....))))..)))))))))....... ( -31.60) >consensus UAUGGGAUUAGUGGCCUCAGACCAUUCGGCCAAUGGCCACGUGCCAGG__ACCCAAGCCAAAGCCCAGG_______CCA_AAG_CCCAGGCCCAAACAGGCUCUGGCCUCAAAACUGA ..((((....((((((...(.((....)).)...))))))..(((((.........((....))........................((((......)))))))))))))....... (-24.13 = -24.65 + 0.52)

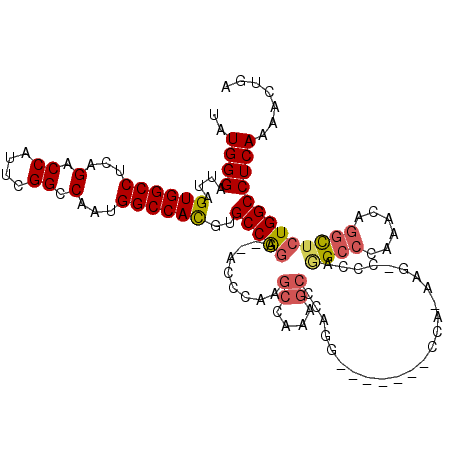

| Location | 10,110,098 – 10,110,207 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -49.35 |
| Consensus MFE | -25.65 |
| Energy contribution | -27.84 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
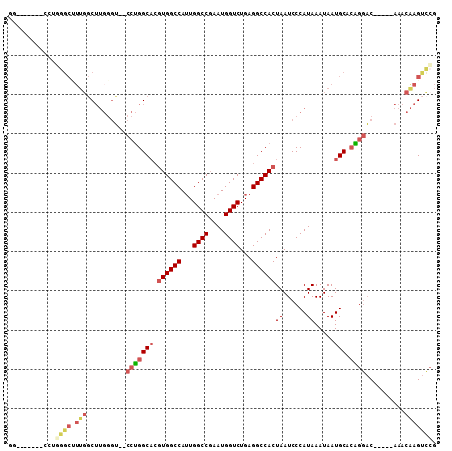
>3L_DroMel_CAF1 10110098 109 - 23771897 UCAGUUUUGAGGCCAGAGCCUCUUUGGGCUUGGG-CUU-GGG-------CCUGGGCUUUGGCUUGGGUGGCCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA .........((((((((((((....((((((...-...-)))-------))))))))))))))).(((((((((((...(((((...)))))....)))..))))))))......... ( -51.30) >DroPse_CAF1 53784 104 - 1 UCAGUUUUGAGGCCAAAAUAAGGUUCGGCCGUGGCACG-UGG-----CAGCGCAGCGUUGCC-----G--GACGGCCAAUGGCCAAUGGCC-AAUGGUCUGAGGCCACUAAUCCCAUA ..........((((............))))(((((...-(((-----(((((...)))))))-----)--...(((((.(((((...))))-).)))))....))))).......... ( -42.40) >DroSec_CAF1 49081 114 - 1 UCAGUUUUGAGGCCAGAGCCUGUUUGGGCCUGGG-CUU-UGGCUUGGGUCCUGGGCUUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA ((((..((.((((((((((((....(((((..((-(..-..)))..))))).)))))))))))).)).--.))))...((((((...((((....))))...)))))).......... ( -60.90) >DroSim_CAF1 50089 95 - 1 UCAGUUUUGAGGCCAGAGCCUGUUUGGGCCUGGG-CCU-UGG-------------------CUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA .........((((....))))((..(((((..((-(..-..)-------------------))..)))--))..))..((((((...((((....))))...)))))).......... ( -43.00) >DroEre_CAF1 53776 114 - 1 UCAGUUUUGAGGCCAGAGCCAGUUUGGGCCUGGA-GCUUGGGCUUGGG-CUUGGGCUUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA ((((..((.(((((((((((((((..((((..(.-..)..))))..))-))..))))))))))).)).--.))))...((((((...((((....))))...)))))).......... ( -57.10) >DroYak_CAF1 51909 102 - 1 UCAGUUUUGAGGCCAGAGCCAGUUUGGGCACGGA-GUUUG-------------GGCCUUGGCUUGGGU--CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA ...(((((.((((((..(((((..(((.((((..-((..(-------------(((((......))))--))..)).)))).))))))))....)))))))))))............. ( -41.40) >consensus UCAGUUUUGAGGCCAGAGCCAGUUUGGGCCUGGG_CCU_GGG_______CCUGGGCUUUGGCUUGGGU__CCUGGCACGUGGCCAUUGGCCGAAUGGUCUGAGGCCACUAAUCCCAUA ...........(((((.(((.(((.((((.........................)))).)))...)))...)))))..((((((...((((....))))...)))))).......... (-25.65 = -27.84 + 2.20)

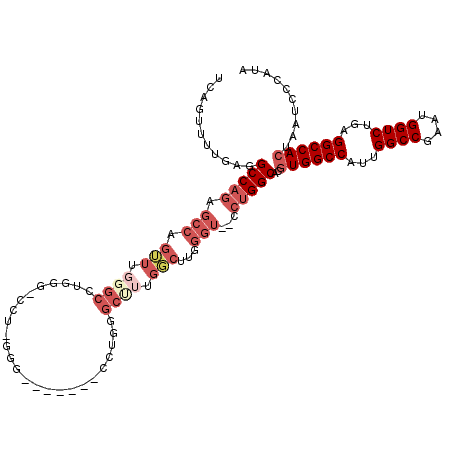

| Location | 10,110,136 – 10,110,240 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.17 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -13.02 |
| Energy contribution | -14.86 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10110136 104 - 23771897 ----ACGAUGCAUA---GGACGGGACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCUCUUUGGGCUUGGG-CUU-GGG-------CCUGGGCUUUGGCUUGGGUGGCCUGGCACGU ----(((.(((...---...(((((((..((((....)))).)))))))(((((((((((.....(((((..((-(..-..)-------))..))))).)))))...)))))).)))))) ( -41.10) >DroPse_CAF1 53821 107 - 1 GCAAAGGCCGCAGAGACGGGGAAAAUUUCAUUGAACCUAAUCAGUUUUGAGGCCAAAAUAAGGUUCGGCCGUGGCACG-UGG-----CAGCGCAGCGUUGCC-----G--GACGGCCAAU .....((((...((((.........))))...((((((.....((((((....)))))).)))))))))).((((.((-(((-----(((((...)))))))-----.--.))))))).. ( -37.20) >DroSec_CAF1 49119 109 - 1 ----ACGAUGCAUA---GGACAGGACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCUGUUUGGGCCUGGG-CUU-UGGCUUGGGUCCUGGGCUUUGGCUUGGGU--CCUGGCACGU ----(((.(((.((---((((((((((..((((....)))).)))))).((((((((((((....(((((..((-(..-..)))..))))).))))))))))))..))--)))))))))) ( -53.50) >DroSim_CAF1 50127 90 - 1 ----ACUAUGCAUA---GGACAGGACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCUGUUUGGGCCUGGG-CCU-UGG-------------------CUUGGGU--CCUGGCACGU ----....(((.((---((((.((.(((((((((......))))...))))))).(((((.....((((....)-)))-.))-------------------)))..))--)))))))... ( -33.40) >DroEre_CAF1 53814 109 - 1 ----ACGAGGCAAG---GGACAGCACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCAGUUUGGGCCUGGA-GCUUGGGCUUGGG-CUUGGGCUUUGGCUUGGGU--CCUGGCACGU ----(((..((.((---(..(((.......)))..)))...(((..((.(((((((((((((((..((((..(.-..)..))))..))-))..))))))))))).)).--.))))).))) ( -46.50) >DroYak_CAF1 51947 97 - 1 ----ACGAAGCCUA---GGACAGGACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCAGUUUGGGCACGGA-GUUUG-------------GGCCUUGGCUUGGGU--CCUGGCACGU ----.....(((.(---((((.((.(((((((((......))))...))))))).(((((((((..(((.....-)))..-------------)))..))))))..))--)))))).... ( -36.00) >consensus ____ACGAUGCAUA___GGACAGGACUUCAUUGAACCUAAUCAGUUUUGAGGCCAGAGCCAGUUUGGGCCUGGG_CCU_GGG_______CCUGGGCUUUGGCUUGGGU__CCUGGCACGU ....................(((((((..((((....)))).)))))))..(((((.(((.(((.((((.........................)))).)))...)))...))))).... (-13.02 = -14.86 + 1.84)

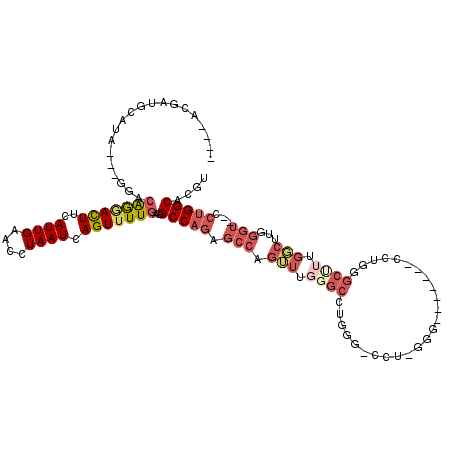

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:21 2006