| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,106,146 – 10,106,244 |
| Length | 98 |
| Max. P | 0.768443 |

| Location | 10,106,146 – 10,106,244 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -13.02 |
| Energy contribution | -12.62 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
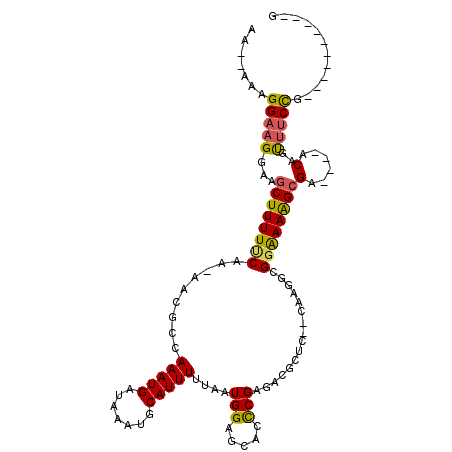
>3L_DroMel_CAF1 10106146 98 + 23771897 AAAGAAAGGAAGGAAGCUUUUUCAA-AACGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACGCUC--CAAGGCGGAAAAGCGA----GCAUUUUCCG----------G .......((((((..(((((((...-..(((((((((.......)))))....((((((.........))))--)).)))))))))))..----...)))))).----------. ( -27.00) >DroPse_CAF1 49685 112 + 1 AA---AAGGAAGGAAGCUUUUCCAAAAAUGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACACGCCGCAAAAUGGGAAGGCGCUUCCACAGCUUCCAAAACGCGAACG ..---......(((((((.((((((((((((............)))))))...))))).....(.((..((((............))))..)))..)))))))............ ( -26.10) >DroSim_CAF1 46049 98 + 1 AAAGAAAGGAAGGAAGCUUUUUCAA-AACGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACGCUC--CAAGGCGGAAAAGCGA----GCAUUUUCCG----------G .......((((((..(((((((...-..(((((((((.......)))))....((((((.........))))--)).)))))))))))..----...)))))).----------. ( -27.00) >DroEre_CAF1 49845 98 + 1 AAAGAAAGGAAGGAAGCUUUUUCAA-AACGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACGCUC--CGUGGCGGAAAAGCGA----ACAUUUUCCG----------G .......((((((..(((((((...-..(((((((((.......)))))...(((((((.........))))--))))))))))))))..----...)))))).----------. ( -27.40) >DroWil_CAF1 142171 82 + 1 ----ACAGGAAGGAAGCUUUUU--A-AAUGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCGCUCGAGACGC------------AAAAGAGA----GCAGUCCCUG----------U ----(((((......(((((..--(-(((((............)))))).....)))))((((.......------------......))----))....))))----------) ( -19.32) >DroPer_CAF1 50028 112 + 1 AA---AAGGAAGGAAGCUUUUCCAAAAAUGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACACGCCGCAAAAUGGGAAGGCGCUUCCACAGCUUCCAAAACGCGAACG ..---......(((((((.((((((((((((............)))))))...))))).....(.((..((((............))))..)))..)))))))............ ( -26.10) >consensus AA__AAAGGAAGGAAGCUUUUUCAA_AACGCCAAAUGAUAAAUGCAUUUUUAAUGGAGCACCCGAGACGCUC__CAAGGCGGAAAAGCGA____ACAGUUUCCG__________G .......(((((...((((((((.........(((((.......)))))....(((.....)))................))))))))(......)..)))))............ (-13.02 = -12.62 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:14 2006