| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,076,614 – 10,076,705 |
| Length | 91 |
| Max. P | 0.802043 |

| Location | 10,076,614 – 10,076,705 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
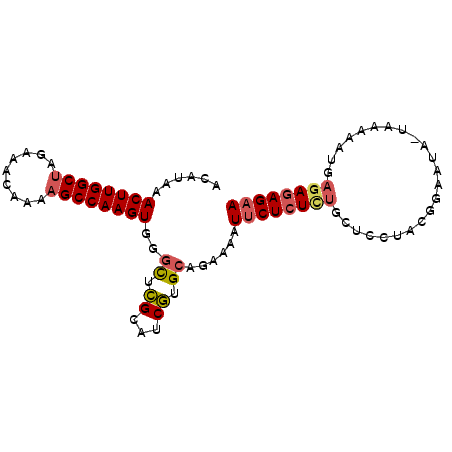
>3L_DroMel_CAF1 10076614 91 + 23771897 ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGCUCGCAUCGUGCAGUAAAUUCUCUCUGCUUCUACGAAAUUUUUAAAAUGAGAGAGAA ......((((((((.........))))))))..(((.((.....)))))...((((((((.(....................))))))))) ( -21.35) >DroSec_CAF1 15871 90 + 1 ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGCUCGCAUCGUGCAGAAAAUUCUCUCUGCUCCUACGGAAUG-UAAAAAUGAGAGAGAA ......((((((((.........))))))))...(((...(((((((((........)))))(((...)))...-.....))))..))).. ( -23.30) >DroSim_CAF1 16235 90 + 1 ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGCUCGCAUCGUGCAGAAAAUUCUCUCUGCUCCUACGGAAUG-UAAAAAUGAGAGAGAA ......((((((((.........))))))))...(((...(((((((((........)))))(((...)))...-.....))))..))).. ( -23.30) >DroEre_CAF1 16529 90 + 1 ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGCUCGCAUCGUGCUGAAAAUUCUCUUUUCUGCUACGGCAUA-UAAAAACAAGAGAGAA ......((((((((.........)))))))).(((.((...)).))).....(((((((((.(((....)))..-.......))))))))) ( -23.60) >DroYak_CAF1 17341 90 + 1 ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGUUCGCAUCGUGCAGAAAAUGCUCUCUUUUGCCACAGAAUA-UAAAAAUAAGAGAGAA ......((((((((.........))))))))...(((((.....)).)))....(((((((.............-.......))))))).. ( -19.55) >DroAna_CAF1 15552 71 + 1 ACAUAAACUUGGCAGAAAACAAAAGCCAAGUGGGUUUGCCUCAUGA--AAAUUUCUCUCU-------------A-UAAA-AU---AAAGAA .(((..(((((((...........)))))))(((....))).))).--............-------------.-....-..---...... ( -11.90) >consensus ACAUAAACUUGGCUAGAAACAAAAGCCAAGUGGGCUCGCAUCGUGCAGAAAAUUCUCUCUGCUCCUACGGAAUA_UAAAAAUGAGAGAGAA ......((((((((.........))))))))..((.((...)).))......((((((((.......................)))))))) (-14.94 = -15.33 + 0.39)

| Location | 10,076,614 – 10,076,705 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -14.19 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
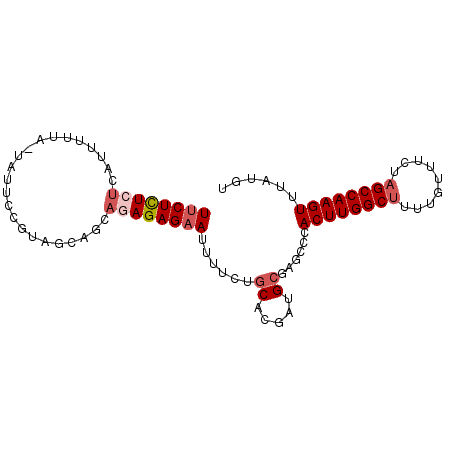
>3L_DroMel_CAF1 10076614 91 - 23771897 UUCUCUCUCAUUUUAAAAAUUUCGUAGAAGCAGAGAGAAUUUACUGCACGAUGCGAGCCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU ((((((((..(((((.........)))))..)))))))).....(((.....))).....((((((((.........))))))))...... ( -21.80) >DroSec_CAF1 15871 90 - 1 UUCUCUCUCAUUUUUA-CAUUCCGUAGGAGCAGAGAGAAUUUUCUGCACGAUGCGAGCCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU ((((((((..((((((-(.....))))))).)))))))).....(((.....))).....((((((((.........))))))))...... ( -25.00) >DroSim_CAF1 16235 90 - 1 UUCUCUCUCAUUUUUA-CAUUCCGUAGGAGCAGAGAGAAUUUUCUGCACGAUGCGAGCCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU ((((((((..((((((-(.....))))))).)))))))).....(((.....))).....((((((((.........))))))))...... ( -25.00) >DroEre_CAF1 16529 90 - 1 UUCUCUCUUGUUUUUA-UAUGCCGUAGCAGAAAAGAGAAUUUUCAGCACGAUGCGAGCCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU ................-..((((((.((.(((((.....))))).)))))..))).....((((((((.........))))))))...... ( -18.80) >DroYak_CAF1 17341 90 - 1 UUCUCUCUUAUUUUUA-UAUUCUGUGGCAAAAGAGAGCAUUUUCUGCACGAUGCGAACCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU (((((((((.((.(((-(.....)))).))))))))(((((........))))))))...((((((((.........))))))))...... ( -19.30) >DroAna_CAF1 15552 71 - 1 UUCUUU---AU-UUUA-U-------------AGAGAGAAAUUU--UCAUGAGGCAAACCCACUUGGCUUUUGUUUUCUGCCAAGUUUAUGU ((((((---((-...)-)-------------))))))......--.(((((((.....))(((((((...........)))))))))))). ( -15.20) >consensus UUCUCUCUCAUUUUUA_UAUUCCGUAGCAGCAGAGAGAAUUUUCUGCACGAUGCGAGCCCACUUGGCUUUUGUUUCUAGCCAAGUUUAUGU ((((((((.......................))))))))......((.....))......((((((((.........))))))))...... (-14.19 = -15.05 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:03 2006