| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 993,232 – 993,334 |
| Length | 102 |
| Max. P | 0.934851 |

| Location | 993,232 – 993,334 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
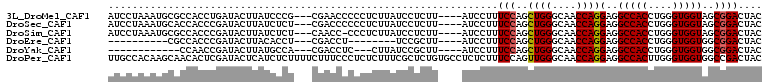
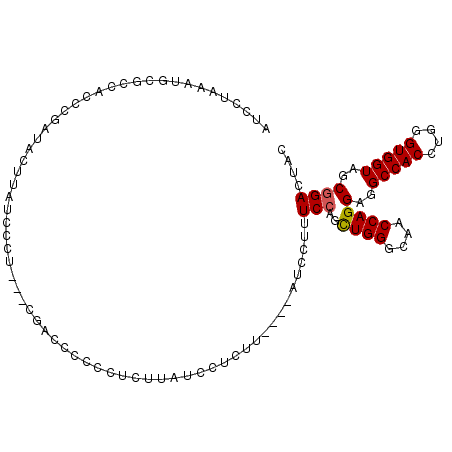
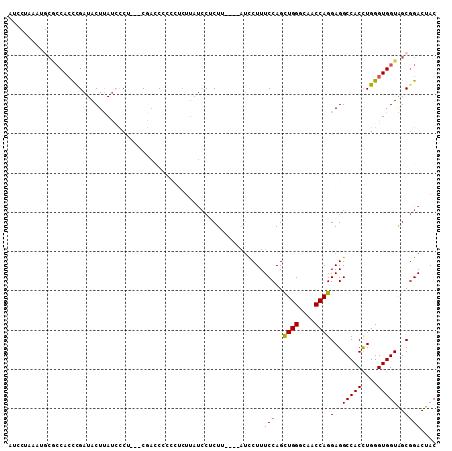
>3L_DroMel_CAF1 993232 102 - 23771897 AUCCUAAAUGCGCCACCUGAUACUUAUCCCG---CGAACCCCCUCUUAUCCUCUU----AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUAGCGGACUAC ............................(((---(..((((((......((((..----...((....))((((....))))))))......))).))).))))..... ( -28.30) >DroSec_CAF1 11504 102 - 1 AUCCUAAAUGCACCACCCGAUACUUAUCUCU---CGACCCCCCUCUUAUCCUCUU----AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUAGCGGACUAC ........(((((((((((((....)))...---.......((((.((......)----)..........((((....))))))))......))))))).)))...... ( -27.80) >DroSim_CAF1 11676 101 - 1 AUCCUAAAUGCGCCACCCGAUACUUAUCUCU---CAACC-CCCUCUUAUCCUCUU----AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUAGCGGACUAC ........(((((((((((((....)))...---.....-.((((.((......)----)..........((((....))))))))......))))))).)))...... ( -27.40) >DroEre_CAF1 11076 84 - 1 ----------CGCCACCCGAUACUUACACCU---CGACCU--------UCCGCUU----AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUGGCGGACUAC ----------((((((((..........(((---((....--------..)....----...........((((....))))))))((....))).)))))))...... ( -27.80) >DroYak_CAF1 12088 87 - 1 ------------CCAACCGAUACUUAUGCCA---CGACCUC---CUUAUCCGCUU----AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUGGCGGACUAC ------------..............(((((---(..((((---(......(((.----.........)))(((....)))))))).(((....)))))))))...... ( -27.80) >DroPer_CAF1 26377 109 - 1 UUGCCACAAGCAACACUCGAUACUCAUCUCUUUUCUUUCCCUCUCUUUCGCUCUGUGCCUCUCUUUCCAGUUGGGCAACCAGGAGGCCACUUGGGUGGUGGCCGACUAC ..(((((...........(((....)))....................(((((.((((((((.........(((....)))))))).)))..))))))))))....... ( -29.00) >consensus AUCCUAAAUGCGCCACCCGAUACUUAUCCCU___CGACCCCCCUCUUAUCCUCUU____AUCCUUUCCAGCUGGGCAACCAGGAGGCCACCUGGGUGGUAGCGGACUAC .................................................................(((..((((....))))(..(((((....)))))..)))).... (-19.06 = -19.08 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:39 2006