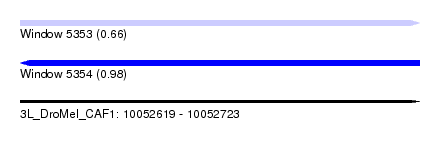
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,052,619 – 10,052,723 |
| Length | 104 |
| Max. P | 0.975419 |

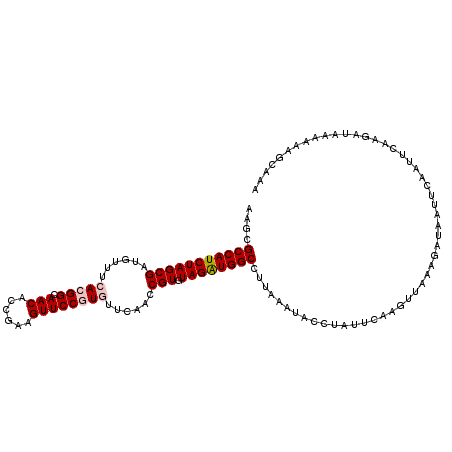
| Location | 10,052,619 – 10,052,723 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10052619 104 + 23771897 ----------UGUAUUGAAUUGAAUUAUUUUUAACUUGAAUAUGUAUUUAAGGCCAUCUACACGGUUGAAUACGGAACUUCGAUGUUGCCGUGAAACAUCGCUAGAUGGCGCUA ----------........................(((((((....)))))))((((((((..((((....((((((((......))).)))))....)))).)))))))).... ( -27.20) >DroSec_CAF1 97383 114 + 1 UGUGCUCAAUUUUCUUGAAUUGAAUUAUCUUUAACUUGUAAAGGUAUUUAAGGCCAUCUACACGGUUGAACACGGAACUUCGCUGUUGCCGUGAAACAUCGCUAGAUGGCGCUU ...(((((((((....)))))))..((((((((.....))))))))......((((((((..((((....((((((((......))).)))))....)))).)))))))))).. ( -34.80) >DroSim_CAF1 107365 114 + 1 UGUGCUCAUUUAUCUUGAAUUGAAUUACCUUUAACCUGUAAAGGUAUUUAAGGCCAUCUACACGGUUGAACACGGAACUUCGCUGUUGCCGUGAAACAUCGCUAGAUGGCGCUU ...((........((((((......((((((((.....))))))))))))))((((((((..((((....((((((((......))).)))))....)))).)))))))))).. ( -35.40) >DroEre_CAF1 101883 107 + 1 UUUCCUUUCUUAUCUGCAAUUGAAUUAAAUUGAGUUUGAACGG-------UGGCCACCUACACGGUUGAAUAAGGAACUUCCGUGUUGCCGUGAAACACCGCUAGAUGGCGCUU ...((.(((....((.(((((......)))))))...))).))-------..((((.(((((((((..((((.((.....)).)))))))))).........))).)))).... ( -24.80) >DroYak_CAF1 102362 114 + 1 UUUCCGUUUUUAUUUUUAAUUGAAUUAUCUUGAACUUGAAUAGGUAUUUGAGGCCACCUAUACGGUUGAACAAGGAACUUGUGUGUUGCCAUGAAACACCGCUAGAUGGCGCAU ..................................((..(((....)))..))((((.(((..((((....((.(((((......))).)).))....)))).))).)))).... ( -20.70) >consensus UGUCCUCAUUUAUCUUGAAUUGAAUUAUCUUUAACUUGAAAAGGUAUUUAAGGCCAUCUACACGGUUGAACACGGAACUUCGCUGUUGCCGUGAAACAUCGCUAGAUGGCGCUU ....................................................((((((((..((((....((((((((......))).)))))....)))).)))))))).... (-21.70 = -22.02 + 0.32)

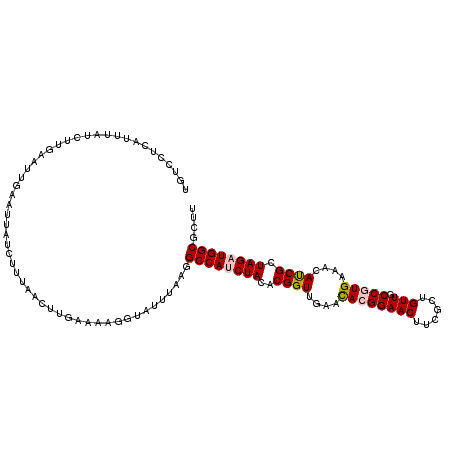
| Location | 10,052,619 – 10,052,723 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10052619 104 - 23771897 UAGCGCCAUCUAGCGAUGUUUCACGGCAACAUCGAAGUUCCGUAUUCAACCGUGUAGAUGGCCUUAAAUACAUAUUCAAGUUAAAAAUAAUUCAAUUCAAUACA---------- .((.(((((((((((.((..(.((((.(((......))))))))..))..))).))))))))))........................................---------- ( -21.10) >DroSec_CAF1 97383 114 - 1 AAGCGCCAUCUAGCGAUGUUUCACGGCAACAGCGAAGUUCCGUGUUCAACCGUGUAGAUGGCCUUAAAUACCUUUACAAGUUAAAGAUAAUUCAAUUCAAGAAAAUUGAGCACA ..(((((((((((((.((...(((((.(((......))))))))..))..))).)))))))).........(((((.....))))).....((((((......))))))))... ( -30.60) >DroSim_CAF1 107365 114 - 1 AAGCGCCAUCUAGCGAUGUUUCACGGCAACAGCGAAGUUCCGUGUUCAACCGUGUAGAUGGCCUUAAAUACCUUUACAGGUUAAAGGUAAUUCAAUUCAAGAUAAAUGAGCACA ..(((((((((((((.((...(((((.(((......))))))))..))..))).))))))))......((((((((.....))))))))....................))... ( -34.30) >DroEre_CAF1 101883 107 - 1 AAGCGCCAUCUAGCGGUGUUUCACGGCAACACGGAAGUUCCUUAUUCAACCGUGUAGGUGGCCA-------CCGUUCAAACUCAAUUUAAUUCAAUUGCAGAUAAGAAAGGAAA ..(.(((((((.((.(((...))).)).((((((..(.........)..)))))))))))))).-------((.(((...(((((((......))))).))....))).))... ( -28.40) >DroYak_CAF1 102362 114 - 1 AUGCGCCAUCUAGCGGUGUUUCAUGGCAACACACAAGUUCCUUGUUCAACCGUAUAGGUGGCCUCAAAUACCUAUUCAAGUUCAAGAUAAUUCAAUUAAAAAUAAAAACGGAAA ....(((((((((((((....((.((.(((......))))).))....))))).)))))))).................................................... ( -24.30) >consensus AAGCGCCAUCUAGCGAUGUUUCACGGCAACACCGAAGUUCCGUGUUCAACCGUGUAGAUGGCCUUAAAUACCUAUUCAAGUUAAAGAUAAUUCAAUUCAAGAUAAAAAAGCAAA ....(((((((((((......(((((.(((......))))))))......))).)))))))).................................................... (-20.46 = -21.02 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:52 2006