
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,042,691 – 10,042,783 |
| Length | 92 |
| Max. P | 0.660011 |

| Location | 10,042,691 – 10,042,783 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
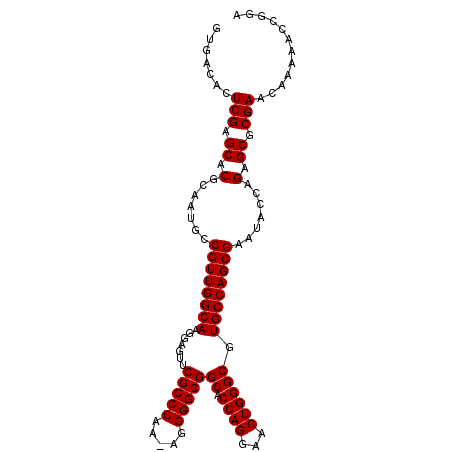
>3L_DroMel_CAF1 10042691 92 + 23771897 ------------UCGCGCUCUGGUAUUCGCUGGCACGCCCAGUUCCUGGUGCCGCCCCUUUUGGGCGAACUCCUUGCCAACGGUAUUGCGUGCUCGAGUGUCAC ------------..((((((.((((((((.(((((.((((((...)))).))(((((.....))))).......))))).)))......))))).))))))... ( -35.40) >DroSec_CAF1 87675 103 + 1 UCCGGUUUUUGUUCGCGCUCUGGUAUUCGCUGGCACGCCCAGUUCCUGGGGCCGCCCCU-UUGGGCGAACUUCUUGCCAACGGCAUUGCGUGCUCGAGUGUCAC ...((..((((..(((((..((....(((.(((((.((((........))))(((((..-..))))).......))))).)))))..)))))..))))..)).. ( -37.40) >DroSim_CAF1 97477 103 + 1 UCCGGUUUUUGUUCGCGCUCUGGUAUUCGCUGGCACGCCCAGUUCCUGGUGCCGCCCCU-UUGGGCGAACUCCUUGCCAACGGCAUUGCGUGCUCGAGUGUCAC ...((..((((..(((((..((....(((.(((((.((((((...)))).))(((((..-..))))).......))))).)))))..)))))..))))..)).. ( -36.30) >consensus UCCGGUUUUUGUUCGCGCUCUGGUAUUCGCUGGCACGCCCAGUUCCUGGUGCCGCCCCU_UUGGGCGAACUCCUUGCCAACGGCAUUGCGUGCUCGAGUGUCAC ..............((((((.((((((((.(((((.((((((...)))).))(((((.....))))).......))))).)))......))))).))))))... (-35.40 = -35.40 + -0.00)



| Location | 10,042,691 – 10,042,783 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -34.57 |
| Energy contribution | -34.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10042691 92 - 23771897 GUGACACUCGAGCACGCAAUACCGUUGGCAAGGAGUUCGCCCAAAAGGGGCGGCACCAGGAACUGGGCGUGCCAGCGAAUACCAGAGCGCGA------------ .......(((.((.(.......((((((((.......(((((.....)))))((.((((...)))))).)))))))).......).)).)))------------ ( -34.04) >DroSec_CAF1 87675 103 - 1 GUGACACUCGAGCACGCAAUGCCGUUGGCAAGAAGUUCGCCCAA-AGGGGCGGCCCCAGGAACUGGGCGUGCCAGCGAAUACCAGAGCGCGAACAAAAACCGGA (((...(((..(((.....)))((((((((.......(((((..-..)))))((((........)))).)))))))).......)))))).............. ( -35.40) >DroSim_CAF1 97477 103 - 1 GUGACACUCGAGCACGCAAUGCCGUUGGCAAGGAGUUCGCCCAA-AGGGGCGGCACCAGGAACUGGGCGUGCCAGCGAAUACCAGAGCGCGAACAAAAACCGGA (((...(((..(((.....)))((((((((.......(((((..-..)))))((.((((...)))))).)))))))).......)))))).............. ( -34.90) >consensus GUGACACUCGAGCACGCAAUGCCGUUGGCAAGGAGUUCGCCCAA_AGGGGCGGCACCAGGAACUGGGCGUGCCAGCGAAUACCAGAGCGCGAACAAAAACCGGA .......(((.((.(.......((((((((.......(((((.....)))))((.((((...)))))).)))))))).......).)).)))............ (-34.57 = -34.57 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:46 2006