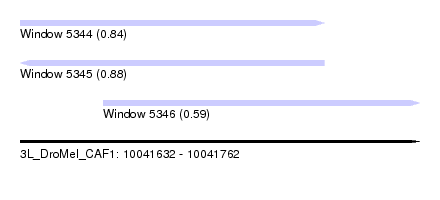
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,041,632 – 10,041,762 |
| Length | 130 |
| Max. P | 0.880738 |

| Location | 10,041,632 – 10,041,731 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
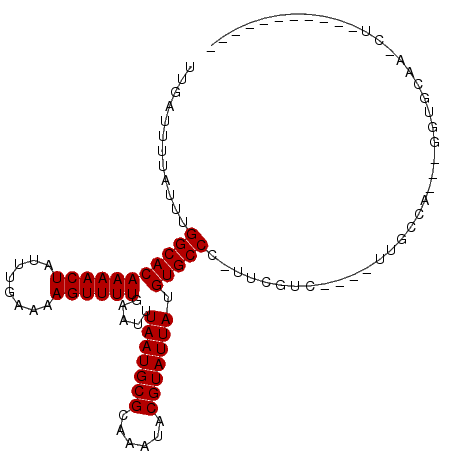
>3L_DroMel_CAF1 10041632 99 + 23771897 ACUAUAU----AU-AUAUACA-AGGGGCAA----GACGAA-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA .......----..-.......-...((((.----......-(.(((.(((.....))).))).)........(((((((........)))))))))))............ ( -15.90) >DroPse_CAF1 92903 95 + 1 -------GCGUAGAGAUCUCC---UGACAG----AUGGAG-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA -------.....((..(((((---......----..))))-)(((((.((((.(((....))).)))).....((((((........)))))))))))........)).. ( -21.70) >DroSim_CAF1 96378 93 + 1 -----------AGCUUGCACA-AGGGGCAA----GACGAA-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA -----------.((((((...-....))))----).)...-.(((((.((((.(((....))).)))).....((((((........)))))))))))............ ( -20.60) >DroEre_CAF1 90983 92 + 1 ------------GCU-GCACGAGGGGGCAA----GACGAA-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA ------------(((-.(.....).)))..----......-.(((((.((((.(((....))).)))).....((((((........)))))))))))............ ( -19.10) >DroAna_CAF1 88435 92 + 1 ---------------UGGAUC---UGGCAAUGAAGAUGCCGAGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCGAAUAGUUUUGUGCCAAAUAAAAUCAA ---------------..(((.---(((((.......))))).(((((.((((.(((....))).)))).....((((((........))))))))))).......))).. ( -20.80) >DroPer_CAF1 92824 95 + 1 -------GCGUAGAGAUCUCC---UGACAG----AUGGAG-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA -------.....((..(((((---......----..))))-)(((((.((((.(((....))).)))).....((((((........)))))))))))........)).. ( -21.70) >consensus ___________AG_GUGCACC___GGGCAA____GACGAA_GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUUUGUGCCAAAUAAAAUCAA ..........................................(((((.((((.(((....))).)))).....((((((........)))))))))))............ (-15.13 = -15.13 + 0.00)


| Location | 10,041,632 – 10,041,731 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10041632 99 - 23771897 UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC-UUCGUC----UUGCCCCU-UGUAUAU-AU----AUAUAGU ............(((((((((((........)))))).....(((((((......))))))).))))).-......----.......(-(((((..-..----)))))). ( -17.00) >DroPse_CAF1 92903 95 - 1 UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC-CUCCAU----CUGUCA---GGAGAUCUCUACGC------- ..(((.......(((((((((((........)))))).....(((((((......))))))).))))).-((((..----......---))))))).......------- ( -21.70) >DroSim_CAF1 96378 93 - 1 UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC-UUCGUC----UUGCCCCU-UGUGCAAGCU----------- ............(((((((((((........)))))).....(((((((......))))))).))))).-...(.(----((((....-...)))))).----------- ( -21.00) >DroEre_CAF1 90983 92 - 1 UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC-UUCGUC----UUGCCCCCUCGUGC-AGC------------ ............(((((((((((........)))))).....(((((((......))))))).))))).-......----((((........))-)).------------ ( -17.90) >DroAna_CAF1 88435 92 - 1 UUGAUUUUAUUUGGCACAAAACUAUUCGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCUCGGCAUCUUCAUUGCCA---GAUCCA--------------- ..(((((.....((((((.....(((((((....))))))).(((((((......)))))))))))))..((((.......)))))---))))..--------------- ( -25.50) >DroPer_CAF1 92824 95 - 1 UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC-CUCCAU----CUGUCA---GGAGAUCUCUACGC------- ..(((.......(((((((((((........)))))).....(((((((......))))))).))))).-((((..----......---))))))).......------- ( -21.70) >consensus UUGAUUUUAUUUGGCACAAAACUAUUUGAAAAGUUUUGAAUUUAAUGCGCAAAUACGUAUUAUGUGCCC_UUCGUC____UUGCCA___GGUGCAA_CU___________ ............(((((((((((........)))))).....(((((((......))))))).))))).......................................... (-16.57 = -16.57 + -0.00)


| Location | 10,041,659 – 10,041,762 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -17.38 |
| Consensus MFE | -9.84 |
| Energy contribution | -10.62 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10041659 103 + 23771897 GAA-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUU-UGUGCCA--AAUAAAAUCAAAACGAAAUCUACAAGGAAA-UAAAAA-CAA-------CAA ...-.(((((.((((.(((....))).)))).....((((((........)))))-)))))).--..............................-......-...-------... ( -15.10) >DroVir_CAF1 88630 102 + 1 GAG-G--AGCAUAAUGCGUAUUUGCGCAUUAAAUGCAAAACUUGGCAAAUAGUUU-UGUGCCAAAAAUAAAAUCCAAACAAAAUUUAACA-GCAA-CAAAAC-UGA-------AAA ..(-(--((((((((((((....))))))))..))).....((((((.(......-).))))))........))).............((-(...-.....)-)).-------... ( -23.30) >DroPse_CAF1 92926 111 + 1 GAG-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUU-UGUGCCA--AAUAAAAUCAAAAGGAAAUGUACAAGAAAAAUAAAAG-GGAAUUGGUUCAG ...-((((...((((.(((....))).))))(((((....(((((.......(((-(((((..--...................)))))))).....)))))-.))))).)))).. ( -15.50) >DroMoj_CAF1 87501 103 + 1 GAG-A--GGCAUAAUGCGUAUUUGCGCAUUAAAUGCAAAACUUGGCAAAUAGUUUGUGUGCCGAAAAUAAAAUCAAAACAAAAUCUAACA-GAAA-UAAACG-GAA-------AAA .((-(--.(((((((((((....))))))))..))).....((((((.((.....)).))))))...................)))....-....-......-...-------... ( -19.60) >DroAna_CAF1 88454 105 + 1 GCCGAGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCGAAUAGUUU-UGUGCCA--AAUAAAAUCAAAAAAAAAUCCACAAGAAUA-UAAAAAGCAA-------CGA .....(((((.((((.(((....))).)))).....((((((........)))))-)))))).--..............................-..........-------... ( -15.30) >DroPer_CAF1 92847 111 + 1 GAG-GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUU-UGUGCCA--AAUAAAAUCAAAAGGAAAUGUACAAGAAAAAUAAAAG-GGAAUUGGUUCAG ...-((((...((((.(((....))).))))(((((....(((((.......(((-(((((..--...................)))))))).....)))))-.))))).)))).. ( -15.50) >consensus GAG_GGGCACAUAAUACGUAUUUGCGCAUUAAAUUCAAAACUUUUCAAAUAGUUU_UGUGCCA__AAUAAAAUCAAAACAAAAUCUACAAGAAAA_UAAAAG_GAA_______CAA .....((((((((((.(((....))).))))......(((((........))))).))))))...................................................... ( -9.84 = -10.62 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:44 2006