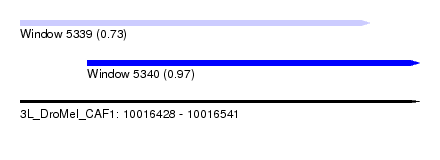
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,016,428 – 10,016,541 |
| Length | 113 |
| Max. P | 0.965769 |

| Location | 10,016,428 – 10,016,527 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -24.09 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10016428 99 + 23771897 CCACCCAGCG-----------------U--UUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCA .......(((-----------------(--((....((((((((...(((((((((((((.((((....))..)))))))))...(....)))))))...))))).)))..)))))). ( -30.70) >DroVir_CAF1 70317 101 + 1 GGCCGGUUUC-----------------CCGCAGGCCGCAAAUGGACCGCAAAUGGCAGCGUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCA ((((.((...-----------------..)).))))((((((((...(((((((((((((((........)))))))).((....))...)))))))...))))).)))......... ( -38.80) >DroPse_CAF1 74565 101 + 1 GUCCCCCCUC-----------------UCGACUGCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCA (((.......-----------------..))).((.(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))..)).....((((....)))) ( -26.80) >DroGri_CAF1 69113 118 + 1 AGCCACUUCCUUUCUCUGGCAUGCAGGCUGCAGGCCACAAAUGGACCGCAAAUGGCAGCAUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCA .((((...........))))((((((((((..(((((....))).))(((((((((((((((........)))))))).((....))...))))))))))))....)))))....... ( -42.00) >DroAna_CAF1 68154 101 + 1 UGCCCCCUCC-----------------UCACUUGCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCA ..........-----------------......((.(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))..)).....((((....)))) ( -26.80) >DroPer_CAF1 74370 101 + 1 GUCCCCCCUC-----------------UCGACUACCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCA (((.......-----------------..)))....(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).........((((....)))) ( -26.20) >consensus GGCCCCCUCC_________________UCGCAUGCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCA ....................................((((((((...(((((((((((((.((((....))..)))))))))...(....)))))))...))))).)))......... (-24.09 = -23.57 + -0.53)



| Location | 10,016,447 – 10,016,541 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10016447 94 + 23771897 AAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUAGAAA-AC------------------------- .(((((...(((((((((((((.((((....))..)))))))))...(....)))))))...)))))(((((....)))))...........-..------------------------- ( -25.10) >DroVir_CAF1 70338 119 + 1 AAAUGGACCGCAAAUGGCAGCGUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUACGUA-GUGUCACACUUAUACCAACACACACUC .(((((...(((((((((((((((........)))))))).((....))...)))))))...)))))(((((....)))))........((.-((((.............)))))).... ( -32.62) >DroGri_CAF1 69151 101 + 1 AAAUGGACCGCAAAUGGCAGCAUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUACAUG-AUGUCACAC------------------ .(((((...(((((((((((((((........)))))))).((....))...)))))))...)))))(((((....)))))...........-.........------------------ ( -30.60) >DroEre_CAF1 71204 94 + 1 AAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUAGAAU-UC------------------------- .(((((...(((((((((((((.((((....))..)))))))))...(....)))))))...)))))(((((....)))))...........-..------------------------- ( -25.10) >DroYak_CAF1 70973 94 + 1 AAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCAUAGAAAUAGAAU-AC------------------------- .(((((...(((((((((((((.((((....))..)))))))))...(....)))))))...)))))(((((....)))))...........-..------------------------- ( -25.10) >DroAna_CAF1 68175 95 + 1 AAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUAGAAAAAC------------------------- .(((((...(((((((((((((.((((....))..)))))))))...(....)))))))...)))))(((((....)))))..............------------------------- ( -25.10) >consensus AAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUAGAAA_AC_________________________ .(((((...(((((((((((((.((((....))..)))))))))...(....)))))))...)))))(((((....)))))....................................... (-24.94 = -24.25 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:38 2006