| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,009,477 – 10,009,575 |
| Length | 98 |
| Max. P | 0.858441 |

| Location | 10,009,477 – 10,009,575 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -31.68 |
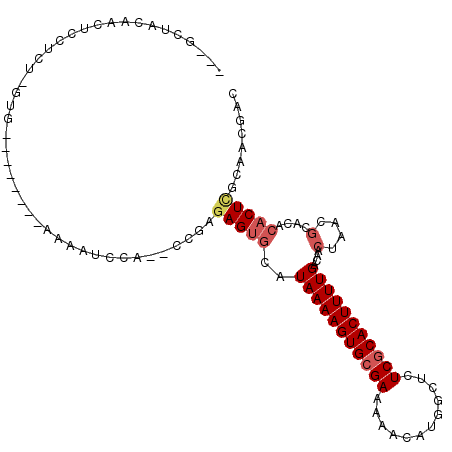
| Consensus MFE | -18.33 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
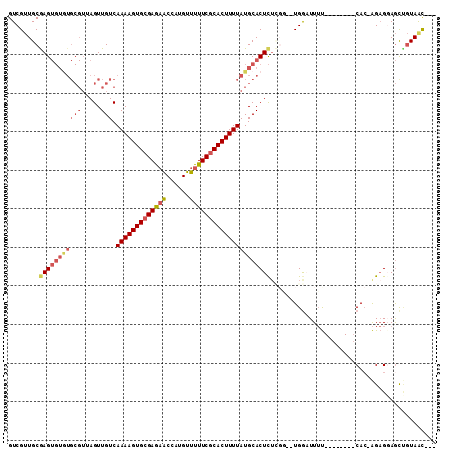
>3L_DroMel_CAF1 10009477 98 + 23771897 GUCGUUGCGAGUGUGUGCGUUAGUUGUCAAAAGUGCGAGAGCCAUGUUUUUCGCACUUUUAUGCACUCUCGG--UGGAUUUU--------CACAAGAGGAGUUGUAGC--- ...(((((((.(..((((((........(((((((((((((......)))))))))))))))))))((((.(--(((....)--------)))..))))).)))))))--- ( -31.80) >DroSec_CAF1 62309 97 + 1 GUCGUUGCGAGUGUGUGCGUUAGUUGUCAAAAGUGCGAGAACCAUGUUUUUCGCACUUUUAUGCACUCUCUG--UGGAUUUU--------CAC-AGAGCAGUUGUAGC--- ...(((((((.(((((((((........(((((((((((((......)))))))))))))))))))..((((--(((....)--------)))-)))))).)))))))--- ( -38.90) >DroSim_CAF1 71981 94 + 1 GUCGUUGCAAGUGUGUGCGUUAGUUGUCAAAAGUGCGAGAACCAUGUUUUUCGCACUUUUAUGCACUC------UGGAUUUU--------CAGAAGAGGAGCUGUAGC--- ...((((((.((..((((((........(((((((((((((......)))))))))))))))))))((------(((....)--------))))......))))))))--- ( -29.80) >DroEre_CAF1 64513 105 + 1 GUCGUUGCGAGUGUGUGCGUUAGUUGUCAAAAGUGCGAGAGCCAUGUUUUUCGCACUUUUAUGCACUCUUGG--UGGCUUUUUUUUUGUGCAC-AGAGCAGCAGUAAU--- ...(((((...((((..((..(((..(((((((((((((((......)))))))))))..........))))--..))).......))..)))-)..)))))......--- ( -35.00) >DroYak_CAF1 63983 97 + 1 GUCGUUGCGAGUGUAUGCGUUAGUUGUCAAAAGUGCGAGAACCAUGUUUUUCGCACUUUUAUGCACUCUGGC--UGAAUUUU--------CAC-AGAGGAGCCGUAAU--- ...((((((((((((((((.....))).(((((((((((((......))))))))))))))))))))).(((--(...(((.--------...-.))).)))))))))--- ( -32.70) >DroAna_CAF1 60518 91 + 1 GUCGCUGCGAG----------AGUUGUCAAAAGUGCGAACGCCAUGUUUUUCUCACUUUUAUGCACUUUCGUCCUGG-UUGC------UGCAC-GG-GGAAUC-UAACAUA ......(((((----------(((.((.(((((((.(((..........))).)))))))..))))))))))(((((-(...------.)).)-))-).....-....... ( -21.90) >consensus GUCGUUGCGAGUGUGUGCGUUAGUUGUCAAAAGUGCGAGAACCAUGUUUUUCGCACUUUUAUGCACUCUCGG__UGGAUUUU________CAC_AGAGGAGCUGUAAC___ ........(((((((((((.....))).(((((((((((((......)))))))))))))))))))))........................................... (-18.33 = -19.55 + 1.22)


| Location | 10,009,477 – 10,009,575 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -13.66 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10009477 98 - 23771897 ---GCUACAACUCCUCUUGUG--------AAAAUCCA--CCGAGAGUGCAUAAAAGUGCGAAAAACAUGGCUCUCGCACUUUUGACAACUAACGCACACACUCGCAACGAC ---((.........(((((((--------......))--.)))))((((..((((((((((............))))))))))(........)))))......))...... ( -21.90) >DroSec_CAF1 62309 97 - 1 ---GCUACAACUGCUCU-GUG--------AAAAUCCA--CAGAGAGUGCAUAAAAGUGCGAAAAACAUGGUUCUCGCACUUUUGACAACUAACGCACACACUCGCAACGAC ---........((((((-(((--------......))--))))(((((..(((((((((((.((......)).)))))))))))....(....)....))))))))..... ( -28.90) >DroSim_CAF1 71981 94 - 1 ---GCUACAGCUCCUCUUCUG--------AAAAUCCA------GAGUGCAUAAAAGUGCGAAAAACAUGGUUCUCGCACUUUUGACAACUAACGCACACACUUGCAACGAC ---......((......((((--------......))------))((((..((((((((((.((......)).))))))))))(........)))))......))...... ( -21.10) >DroEre_CAF1 64513 105 - 1 ---AUUACUGCUGCUCU-GUGCACAAAAAAAAAGCCA--CCAAGAGUGCAUAAAAGUGCGAAAAACAUGGCUCUCGCACUUUUGACAACUAACGCACACACUCGCAACGAC ---.....(((.....(-((((...........((.(--(.....)))).(((((((((((............))))))))))).........))))).....)))..... ( -23.80) >DroYak_CAF1 63983 97 - 1 ---AUUACGGCUCCUCU-GUG--------AAAAUUCA--GCCAGAGUGCAUAAAAGUGCGAAAAACAUGGUUCUCGCACUUUUGACAACUAACGCAUACACUCGCAACGAC ---.....((((.....-((.--------...))..)--))).(((((..(((((((((((.((......)).)))))))))))....(....)....)))))........ ( -23.80) >DroAna_CAF1 60518 91 - 1 UAUGUUA-GAUUCC-CC-GUGCA------GCAA-CCAGGACGAAAGUGCAUAAAAGUGAGAAAAACAUGGCGUUCGCACUUUUGACAACU----------CUCGCAGCGAC ..(((.(-((.(((-..-((...------...)-)..)))(((((((((......((.(........).))....))))))))).....)----------)).)))..... ( -18.20) >consensus ___GCUACAACUCCUCU_GUG________AAAAUCCA__CCGAGAGUGCAUAAAAGUGCGAAAAACAUGGCUCUCGCACUUUUGACAACUAACGCACACACUCGCAACGAC ...........................................(((((..(((((((((((............)))))))))))....(....)....)))))........ (-13.66 = -14.38 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:34 2006