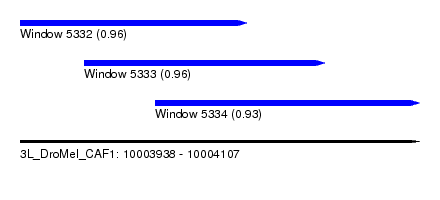
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,003,938 – 10,004,107 |
| Length | 169 |
| Max. P | 0.958656 |

| Location | 10,003,938 – 10,004,034 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10003938 96 + 23771897 UGAGUGGUGCGAUAAAGCAGUCAACAGACAGAAAGGCGCGG--AAGAUGCUACAGAGCGGGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAUUA- ((((((.(((......((.(((....))).....(((((..--..).)))).....)).(((((((........))))))).)))..)))))).....- ( -30.70) >DroSec_CAF1 56621 93 + 1 UGAGUGGUGCGAUAAAGCAGUCAACAGGCAGAAAGGCGCGGGGGGGAUGCU-----GCGGGGUAUCGACUAUUAGAUACCCAGUAUGCAUUAAAAAUA- ....(((((((....((((..(..(..((........))..)..)..))))-----((.(((((((........))))))).)).)))))))......- ( -28.10) >DroSim_CAF1 66291 96 + 1 UGAGUGGUGCGAUAAAGCAGUCAACAGACAGAAAGGCGCGG--AAGAUGCUACAGAGCGGGGUAUCGACUAUUAGAUACCCAGUAAGCAAUCAAAAUA- (((....(((......((.(((....))).....(((((..--..).)))).....)).(((((((........))))))).....))).))).....- ( -27.70) >DroEre_CAF1 58840 96 + 1 UGAGUGGUGCGAUAAAGCAGUCAGCAGACAGAAAGGCGCAG--AAGAUGCUGCAGUGCCAGGCAACGACUAUAAGAUACCCAGUCAGCACUCAAAAUU- ((((((.((((((......))).)))(((.....(((((((--......))))...))).(....)................)))..)))))).....- ( -25.50) >DroYak_CAF1 58092 92 + 1 UGAGUGGUGCGAUAGAGCAGUCGACAGACAGAAAG-----G--AAGAUGCUACAGAGCGGGGUAUCGACUACAAGAUACCCAAUAGGCAUUCAAAAUAU ((((((.(((......)))(((....)))......-----.--....((((....))))(((((((........)))))))......))))))...... ( -25.30) >consensus UGAGUGGUGCGAUAAAGCAGUCAACAGACAGAAAGGCGCGG__AAGAUGCUACAGAGCGGGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAAUA_ ((((((..((.....(((((((....))).........(......).)))).....)).(((((((........)))))))......))))))...... (-19.46 = -20.38 + 0.92)

| Location | 10,003,965 – 10,004,067 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -11.54 |
| Energy contribution | -12.82 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10003965 102 + 23771897 ACAGAAAGGCGCGG--AAGAUGCUACAGAGCGGGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAUUA-----AGUAAGCAGCUUACCAAAAUAGUACACUUAA---AG .......(((((..--..).))))...(((.((((((((........))))))).((((((..........-----........))))))..........).)))..---.. ( -24.37) >DroSec_CAF1 56648 99 + 1 GCAGAAAGGCGCGGGGGGGAUGCU-----GCGGGGUAUCGACUAUUAGAUACCCAGUAUGCAUUAAAAAUA-----UGUAAGCCGCUUACCAAAAUAGUACAUUUAA---AG .......((((((.....((((((-----((.(((((((........))))))).))).))))).......-----)))..))).......................---.. ( -25.80) >DroSim_CAF1 66318 102 + 1 ACAGAAAGGCGCGG--AAGAUGCUACAGAGCGGGGUAUCGACUAUUAGAUACCCAGUAAGCAAUCAAAAUA-----AGUUAGCAGCUUACCAAAAUAGUACAUUUAA---AA .......(((((..--..).))))...(.((.(((((((........))))))).((((((..........-----........)))))).......)).)......---.. ( -23.47) >DroEre_CAF1 58867 105 + 1 ACAGAAAGGCGCAG--AAGAUGCUGCAGUGCCAGGCAACGACUAUAAGAUACCCAGUCAGCACUCAAAAUU-----AGUAAGCCUCCUACCAAAAAAGUACACGCCAUGUAC ......((((((((--......))))(((((..(....)((((...........)))).))))).......-----.....))))............(((((.....))))) ( -23.70) >DroYak_CAF1 58119 100 + 1 ACAGAAAG-----G--AAGAUGCUACAGAGCGGGGUAUCGACUACAAGAUACCCAAUAGGCAUUCAAAAUAUAUCAAAUAAAACUA--GACAAAAUAGUAUAUUUGA---AG ........-----.--..((((((........(((((((........)))))))....)))))).........(((((((..((((--.......)))).)))))))---.. ( -22.00) >consensus ACAGAAAGGCGCGG__AAGAUGCUACAGAGCGGGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAAUA_____AGUAAGCCGCUUACCAAAAUAGUACAUUUAA___AG ..................((((((.....((.(((((((........))))))).)).))))))................................................ (-11.54 = -12.82 + 1.28)



| Location | 10,003,995 – 10,004,107 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -8.80 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10003995 112 + 23771897 GGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAUUA-----AGUAAGCAGCUUACCAAAAUAGUACACUUAA---AGUAUGGUUAGUUACACAAUUUCUAUAGGUUCCUAUGGUGGA (((((((........)))))))..(((((..........-----........)))))(((..((((...(((((.---((..((((.....)).))....)).)))))..))))..))). ( -24.77) >DroSec_CAF1 56675 112 + 1 GGGUAUCGACUAUUAGAUACCCAGUAUGCAUUAAAAAUA-----UGUAAGCCGCUUACCAAAAUAGUACAUUUAA---AGUAUGGUUAGUUACACAAUUUGUAUAGGUUCCUACCGUGGA (((((((........)))))))....(((((.......)-----))))..((((...........(((((.....---.(((........)))......))))).(((....))))))). ( -25.90) >DroSim_CAF1 66348 112 + 1 GGGUAUCGACUAUUAGAUACCCAGUAAGCAAUCAAAAUA-----AGUUAGCAGCUUACCAAAAUAGUACAUUUAA---AAUAUGGUUAGUUAUACAAUUUGUAUAGGUUUCUACCGUGGA (((((((........))))))).((((((..........-----........)))))).................---..((((((.(.((((((.....)))))).)....)))))).. ( -26.87) >DroEre_CAF1 58897 92 + 1 AGGCAACGACUAUAAGAUACCCAGUCAGCACUCAAAAUU-----AGUAAGCCUCCUACCAAAAAAGUACACGCCAUGUACUAAU--------------UUGUAUAAGUUUA--------- ((((...((((...........))))...(((.......-----)))..)))).....((((..((((((.....))))))..)--------------)))..........--------- ( -13.40) >DroYak_CAF1 58144 101 + 1 GGGUAUCGACUACAAGAUACCCAAUAGGCAUUCAAAAUAUAUCAAAUAAAACUA--GACAAAAUAGUAUAUUUGA---AGUAUG--------------GCGCAUAAGUUCAAGUCAAGGA (((((((........)))))))....(((.......((((.(((((((..((((--.......)))).)))))))---.))))(--------------((......)))...)))..... ( -22.90) >consensus GGGUAUCGACUAUAAGAUACCCAGUAAGCAUUCAAAAUA_____AGUAAGCCGCUUACCAAAAUAGUACAUUUAA___AGUAUGGUUAGUUA_ACAAUUUGUAUAGGUUCCUACCGUGGA (((((((........))))))).................................................................................................. ( -8.80 = -9.40 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:32 2006