
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,978,787 – 9,978,897 |
| Length | 110 |
| Max. P | 0.798502 |

| Location | 9,978,787 – 9,978,897 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
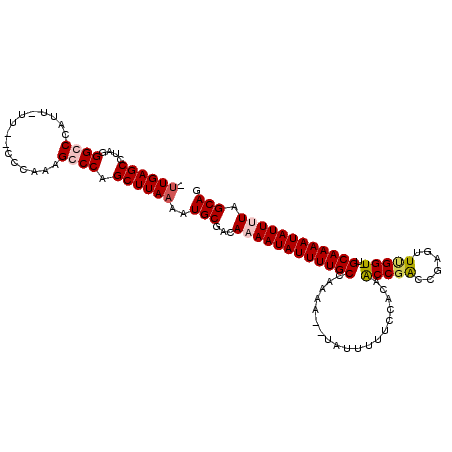
>3L_DroMel_CAF1 9978787 110 + 23771897 --UGAGCCUAGGGCCCAUUUUUCCCCCAAAGCCCAGCUUAAAAUGCGACAUAAUAUUUUGCCUAAA--UAUUUUUCCACAACCGACCGAGUUUGGU-UGCAAAAUAUUUUAGCAG --(((((...((((................)))).)))))..................(((..(((--((((((....(((((((......)))))-)).)))))))))..))). ( -24.29) >DroSec_CAF1 31575 111 + 1 AUUGAGCCUAGGGCCCAUU-UUCCCCCAAAGCCCAGCUUAAAAUGCGACAUAAUAUUUUGCCUAUA--UAUUUUUCCACAACCGACCGAGUUUGGU-UGCAAAAUAUUUUAGCAG .((((((...((((.....-..........)))).))))))..(((.....((((((((((.....--...........((((((......)))))-)))))))))))...))). ( -25.15) >DroEre_CAF1 33771 112 + 1 UUUGAGCCUAGGGCCCAUU-UU--CCCAAAGGCCAGCUUAAAAUGCGGCAAAAUAUUUUGCCAAAACACAUUUUUCCACAACCGACCAAGUUAGGUUUGCAAAAUAUUUUAGCAG (((((((....((((....-..--......)))).))))))).(((...((((((((((((.((((.....))))........((((......)))).)))))))))))).))). ( -26.40) >DroYak_CAF1 31956 109 + 1 CUUGAGCCUAGGGCCCAUU-UU--CCCAAAGGCCAGCUUAAAAUGCGGCAAAAUAUUUUGCCAAAA--UAUUUUUCCACUGCCGGCCGUGUUCGGU-UGCAAAAUAUUUUAGCAG .((((((....((((....-..--......)))).))))))..(((((((((....))))))((((--(((((......(((.(((((....))))-))))))))))))).))). ( -32.70) >consensus _UUGAGCCUAGGGCCCAUU_UU__CCCAAAGCCCAGCUUAAAAUGCGACAAAAUAUUUUGCCAAAA__UAUUUUUCCACAACCGACCGAGUUUGGU_UGCAAAAUAUUUUAGCAG .((((((....((((...............)))).))))))..(((...((((((((((((...................(((((......)))))..)))))))))))).))). (-19.76 = -20.76 + 1.00)

| Location | 9,978,787 – 9,978,897 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -21.83 |
| Energy contribution | -23.07 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9978787 110 - 23771897 CUGCUAAAAUAUUUUGCA-ACCAAACUCGGUCGGUUGUGGAAAAAUA--UUUAGGCAAAAUAUUAUGUCGCAUUUUAAGCUGGGCUUUGGGGGAAAAAUGGGCCCUAGGCUCA-- ..((((((((.(((..((-(((..........)))))..))).....--....((((........))))..))))).))).((((((.((((..........)))))))))).-- ( -29.00) >DroSec_CAF1 31575 111 - 1 CUGCUAAAAUAUUUUGCA-ACCAAACUCGGUCGGUUGUGGAAAAAUA--UAUAGGCAAAAUAUUAUGUCGCAUUUUAAGCUGGGCUUUGGGGGAA-AAUGGGCCCUAGGCUCAAU ..((((((((.(((..((-(((..........)))))..))).....--....((((........))))..))))).)))(((((((.((((...-......))))))))))).. ( -29.90) >DroEre_CAF1 33771 112 - 1 CUGCUAAAAUAUUUUGCAAACCUAACUUGGUCGGUUGUGGAAAAAUGUGUUUUGGCAAAAUAUUUUGCCGCAUUUUAAGCUGGCCUUUGGG--AA-AAUGGGCCCUAGGCUCAAA .(((...........)))..(((((...((((((((.....(((((((.....((((((....))))))))))))).)))))))).)))))--..-..((((((...)))))).. ( -33.60) >DroYak_CAF1 31956 109 - 1 CUGCUAAAAUAUUUUGCA-ACCGAACACGGCCGGCAGUGGAAAAAUA--UUUUGGCAAAAUAUUUUGCCGCAUUUUAAGCUGGCCUUUGGG--AA-AAUGGGCCCUAGGCUCAAG .(((...........)))-.(((((...(((((((.((((.((((((--(((.....))))))))).)))).......)))))))))))).--..-..((((((...)))))).. ( -34.00) >consensus CUGCUAAAAUAUUUUGCA_ACCAAACUCGGUCGGUUGUGGAAAAAUA__UUUAGGCAAAAUAUUAUGCCGCAUUUUAAGCUGGCCUUUGGG__AA_AAUGGGCCCUAGGCUCAA_ ..((((((((.(((..((.(((......)))....))..)))...........((((((....))))))..))))).)))(((((((.(((...........))).))))))).. (-21.83 = -23.07 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:16 2006