
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,974,460 – 9,974,570 |
| Length | 110 |
| Max. P | 0.709408 |

| Location | 9,974,460 – 9,974,570 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.33 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -9.45 |
| Energy contribution | -9.53 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9974460 110 + 23771897 GCCCGCUCACCUU-GG--UUUCC---UCGAGGAAUGGGCCCAUAGAUUGCAGUUUUAUGUCACAACAUUCAAACAAAUAGCCGGCCAUUUAAAACGGUUUAAAAGCCAUUGCUGCA (((((....((((-((--.....---))))))..)))))........((((((...((((....))))..............(((..((((((....)))))).)))...)))))) ( -25.10) >DroVir_CAF1 28484 97 + 1 GUCUGCUAAUCCUGUG--CCCAC-------UAAAUGGA-CUAUAGAUUGCACUUUUAUGUCACAACAUUCAGGCAAAUCGUUGGCCAUUUAAAACGGCUUAAAGGC-A-------- ..........(((..(--((...-------(((((((.-(((..((((((.((...((((....))))..)))).))))..))))))))))....)))....))).-.-------- ( -22.50) >DroPse_CAF1 29519 114 + 1 GCUCGCUCACCUU-GUGGCCAACCGGCCCAGAAAUGGG-CCAUAGAUUGCAGUUUUAUGUCACAACAUUCAAACAAAUCGUUGGCCAUUUAAAACGGUUUAAAAGCCAUUGCUGCA ((..((.......-(((((((((.((((((....))))-))...((((...((((.((((....))))..)))).))))))))))))).......((((....))))...)).)). ( -41.20) >DroWil_CAF1 44078 106 + 1 AUAUGCUUAUGUG-CA--U---A---UACACAAAUGGG-CAAUAGAUUGCAAUUUAACGUCACAACAUUCAAGCAAAUCGUUGGCCAUUUAAAACAGUUUAAAAGCCAUUGCAAUC ...((((((((((-..--.---.---..))))..))))-))...(((((((((..((((...................))))(((..((((((....)))))).)))))))))))) ( -22.81) >DroAna_CAF1 26727 98 + 1 -------CACCUU-UG--CCUCC-------GGAAUGGG-CCAUAGAUUGCAGUUUUAUGUCGCAACAUUCAAACAAAUCGCCGGCCAUUUAAAACGGUUUAAAAGCCAUUGCUGCA -------...(((-(.--...((-------(.....((-((...((((...((((.((((....))))..)))).))))...))))........)))....))))........... ( -19.02) >DroPer_CAF1 29207 114 + 1 GCUCGCUCACCUU-GUGGCCAACCGGCCCAGAAAUGGG-CCAUAGAUUGCAGUUUUAUGUCACAACAUUCAAACAAAUCGUUGGCCAUUUAAAACGGUUUAAAAGCCAUUGCUGCA ((..((.......-(((((((((.((((((....))))-))...((((...((((.((((....))))..)))).))))))))))))).......((((....))))...)).)). ( -41.20) >consensus GCCCGCUCACCUU_GG__CCAAC____CCAGAAAUGGG_CCAUAGAUUGCAGUUUUAUGUCACAACAUUCAAACAAAUCGUUGGCCAUUUAAAACGGUUUAAAAGCCAUUGCUGCA ...............................((((((..((...((((....((..((((....))))..))...))))...)))))))).....((((....))))......... ( -9.45 = -9.53 + 0.09)

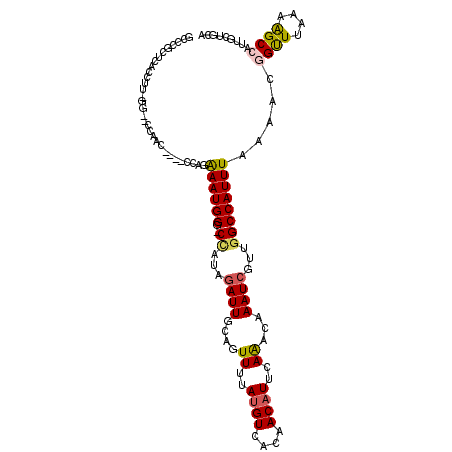
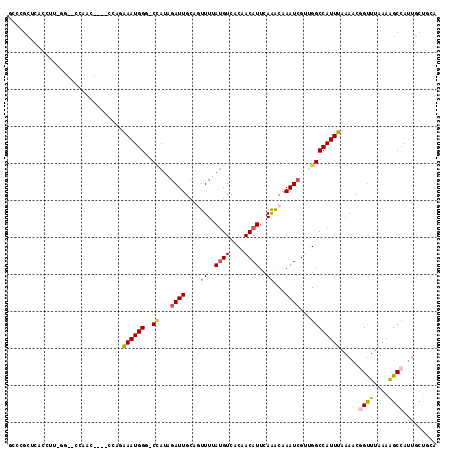
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:14 2006