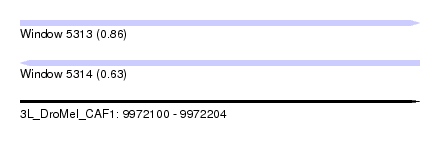
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,972,100 – 9,972,204 |
| Length | 104 |
| Max. P | 0.861439 |

| Location | 9,972,100 – 9,972,204 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -24.74 |
| Energy contribution | -26.02 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9972100 104 + 23771897 UUAAGUGUCUUUCGUUGGCCUUAAAGCCGCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACGGCCAAAUUGGCACUUAUCGAACUGGCGAGAGCCCCAU .((((((((.....((((((.......((((....))))....((((.(((......)))))))..))))))...)))))))).......(((....))).... ( -38.60) >DroSec_CAF1 24851 104 + 1 UUAAGUGUCUUUCGUUGGCCUUAAAGCCGCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACGGCCAAAUUGGCACUUAUCGAACUGGCGAGAGCCCCAU .((((((((.....((((((.......((((....))))....((((.(((......)))))))..))))))...)))))))).......(((....))).... ( -38.60) >DroSim_CAF1 32465 104 + 1 UUAAGUGUCUUUCGUUGGCCUUAAAGCCGCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACGGCCAAAUUGGCACUUAUCGAACUGGCGAGAGCCCCAU .((((((((.....((((((.......((((....))))....((((.(((......)))))))..))))))...)))))))).......(((....))).... ( -38.60) >DroEre_CAF1 25113 104 + 1 UUAAGUGCCUUUCCUUGGCCUUAAAGCCGCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACGGCCAAAUUGGCACUUAUCGAACGGGCGAGAACCCCAU .((((((((.....((((((.......((((....))))....((((.(((......)))))))..))))))...))))))))......(((.(....)))).. ( -37.70) >DroYak_CAF1 24969 104 + 1 UUAAGUGUCUUUCGUUGGACUUAAAGCCUCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACAGCCAAAUUGGCACUUAUCGAACUGUCGAUAACCCGAU ...........(((.(((.((((((((((((..((((....))))))))))))))).).))).....(((.....)))..(((((((....)))))))..))). ( -31.70) >DroAna_CAF1 24441 82 + 1 AUAAGUGCCUCCAGCUGGCAUUAAAGUCGCACU----CGAACUCCUA-AACUUUAAUGCCCAUUAUGGCCAAAUAGCCACUUAUCGG----------------- (((((((.(((((...(((((((((((......----..........-.))))))))))).....)))......)).)))))))...----------------- ( -19.93) >consensus UUAAGUGUCUUUCGUUGGCCUUAAAGCCGCAUUUCUGCGAACGGAUGAGGCUUUAAUGCCCAUUACGGCCAAAUUGGCACUUAUCGAACUGGCGAGAGCCCCAU .((((((((.....((((((((((((((.(((((........))))).))))))))..........))))))...))))))))..................... (-24.74 = -26.02 + 1.28)

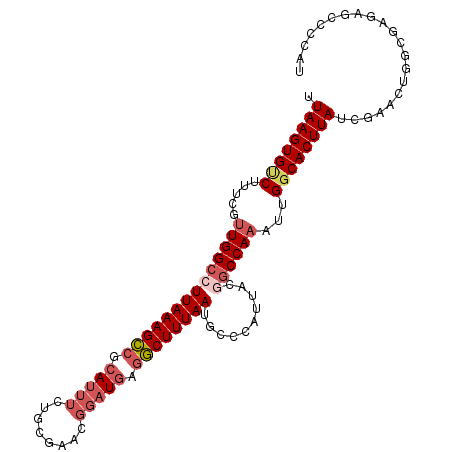
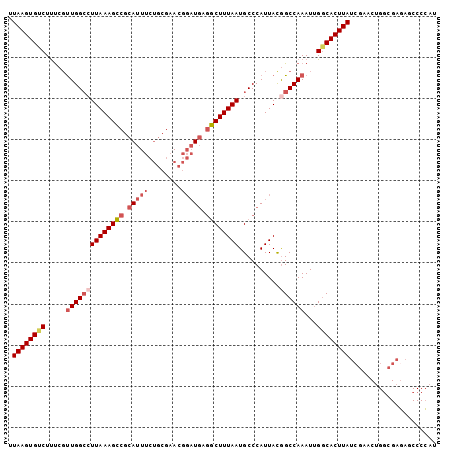
| Location | 9,972,100 – 9,972,204 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9972100 104 - 23771897 AUGGGGCUCUCGCCAGUUCGAUAAGUGCCAAUUUGGCCGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGCGGCUUUAAGGCCAACGAAAGACACUUAA ....(((....))).......((((((.....((((((...((((((......))).)))....(((((....)))))......))))))(....).)))))). ( -33.90) >DroSec_CAF1 24851 104 - 1 AUGGGGCUCUCGCCAGUUCGAUAAGUGCCAAUUUGGCCGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGCGGCUUUAAGGCCAACGAAAGACACUUAA ....(((....))).......((((((.....((((((...((((((......))).)))....(((((....)))))......))))))(....).)))))). ( -33.90) >DroSim_CAF1 32465 104 - 1 AUGGGGCUCUCGCCAGUUCGAUAAGUGCCAAUUUGGCCGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGCGGCUUUAAGGCCAACGAAAGACACUUAA ....(((....))).......((((((.....((((((...((((((......))).)))....(((((....)))))......))))))(....).)))))). ( -33.90) >DroEre_CAF1 25113 104 - 1 AUGGGGUUCUCGCCCGUUCGAUAAGUGCCAAUUUGGCCGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGCGGCUUUAAGGCCAAGGAAAGGCACUUAA ...((((....))))......((((((((..(((((((...((((((......))).)))....(((((....)))))......)))))))....)))))))). ( -42.00) >DroYak_CAF1 24969 104 - 1 AUCGGGUUAUCGACAGUUCGAUAAGUGCCAAUUUGGCUGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGAGGCUUUAAGUCCAACGAAAGACACUUAA .((((.(((((((....))))))).)(((.....))).....(((((.(((((((((((((.(....).))..)))))))))))))))).)))........... ( -34.90) >DroAna_CAF1 24441 82 - 1 -----------------CCGAUAAGUGGCUAUUUGGCCAUAAUGGGCAUUAAAGUU-UAGGAGUUCG----AGUGCGACUUUAAUGCCAGCUGGAGGCACUUAU -----------------...(((((((.((..(..(((.....)((((((((((((-.....((...----...)))))))))))))).))..))).))))))) ( -25.70) >consensus AUGGGGCUCUCGCCAGUUCGAUAAGUGCCAAUUUGGCCGUAAUGGGCAUUAAAGCCUCAUCCGUUCGCAGAAAUGCGGCUUUAAGGCCAACGAAAGACACUUAA ....(((....))).......((((((.(...((((((...((((((......))).)))....(((((....)))))......)))))).....).)))))). (-22.18 = -22.50 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:12 2006