
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,968,287 – 9,968,468 |
| Length | 181 |
| Max. P | 0.995013 |

| Location | 9,968,287 – 9,968,395 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9968287 108 - 23771897 CGAUUAGUGAUGCAGC--UAGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUAAAUUUCGGAUAACUCGCAAU .(((.(((((((((..--...))))))))))))(((((((((....(((((((((..(((........)))))))).)))......)....))))))))).......... ( -24.70) >DroSec_CAF1 21089 110 - 1 CGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUGGAUUAAAUUCGGAUUACUCGCAAU .(((.((((((((((.....)))))))))))))(((((((((((((.((((((((..(((........)))))))).)))...))))))....))))))).......... ( -28.20) >DroSim_CAF1 28670 110 - 1 CGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGGAUUACUCGCAAU .(((.((((((((((.....)))))))))))))(((((((((....(((((((((..(((........)))))))).)))......)....))))))))).......... ( -26.60) >DroEre_CAF1 21309 108 - 1 CGAUUAGUGAUGCAUC--UAGUGCAUCAUUAUACCAUGAAAUACAGCACCUUUUUGGCGCACAUAAAAGUGAAAAAUGGUAAAUUAGAUUAAUUUCGCAUUACUCGCAAU .((.(((((((((((.--..)))))))))))((((((........((.((.....)).)).(((....)))....)))))).............))((.......))... ( -25.00) >DroYak_CAF1 21112 108 - 1 CGAUUAGUGAUGCAUC--UACUGCAUCAUUAUUACAUGACAUACAGCACCUUUUUGGCGCACAUCAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGAAUUACCAGCAAU ....((((((((((..--...))))))))))..............((.((.((((((......)))))).))....((((((....((......))...))))))))... ( -24.80) >consensus CGAUUAGUGAUGCAUC__UAGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGGAUUACUCGCAAU ....(((((((((((.....)))))))))))..............((((((((((.((..........)).))))).))).((((.((......)).))))....))... (-21.70 = -21.74 + 0.04)

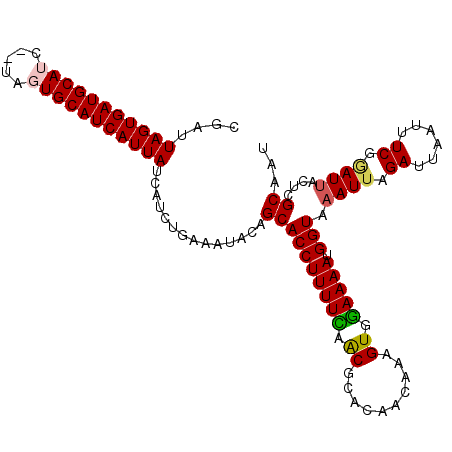

| Location | 9,968,318 – 9,968,434 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -20.59 |
| Energy contribution | -21.77 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9968318 116 + 23771897 CCAUUUUCCACUUUGUUGUGCGUUGAAAAGGUGCUGUAUUUCAGAUGAUAAUGAUGCACUA--GCUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUCCUGUGCCUAUUUAAAUAUAUA ........(((......)))..(((((.(((..(..........(((((..(((((((...--..)))))))...)))))((((......)))-).....)..))).)))))....... ( -21.20) >DroSec_CAF1 21120 118 + 1 CCAUUUUCCACUUUGUUGUGCGUUGAAAAGGUGCUGAAUUUCAGAUGAUAAUGAUGCACUGGAGAUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUGCUGUGCCUAUUUAAAUAUAUA ........(((......)))..(((((.(((..(..........(((((..((((((((....).)))))))...)))))...((((((....-)))))))..))).)))))....... ( -26.80) >DroSim_CAF1 28701 118 + 1 CCAUUUUCCACUUUGUUGUGCGUUGAAAAGGUGCUGCAUUUCAGAUGAUAAUGAUGCACUAGAGAUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUGCUGUGCCUAUUUAAAUAUAUA ........(((......)))..(((((.(((..(..........(((((..((((((((....).)))))))...)))))...((((((....-)))))))..))).)))))....... ( -26.80) >DroYak_CAF1 21143 117 + 1 CCAUUUUCCACUUUGAUGUGCGCCAAAAAGGUGCUGUAUGUCAUGUAAUAAUGAUGCAGUA--GAUGCAUCACUAAUCGUAUUAGCCAAAUACAUUGGCUGUUCCCAUUAAAAUACAUA ..............((((((((((.....)))))...)))))(((((....(((((((...--..))))))).((((.(...(((((((.....)))))))...).))))...))))). ( -29.70) >consensus CCAUUUUCCACUUUGUUGUGCGUUGAAAAGGUGCUGUAUUUCAGAUGAUAAUGAUGCACUA__GAUGCAUCACUAAUCGUAUUAGCAAAAUAA_UUUGCUGUGCCUAUUUAAAUAUAUA ........(((......)))..(((((.((((((..........(((((..(((((((.......)))))))...)))))...((((((.....)))))))))))).)))))....... (-20.59 = -21.77 + 1.19)

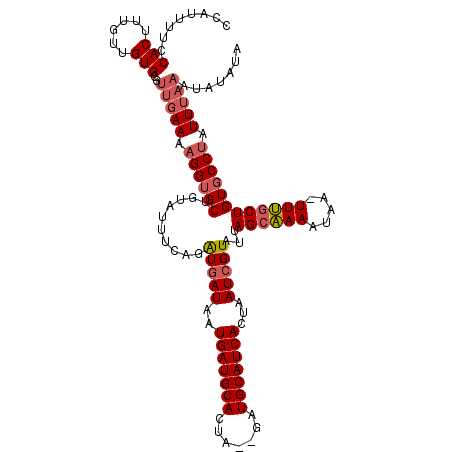
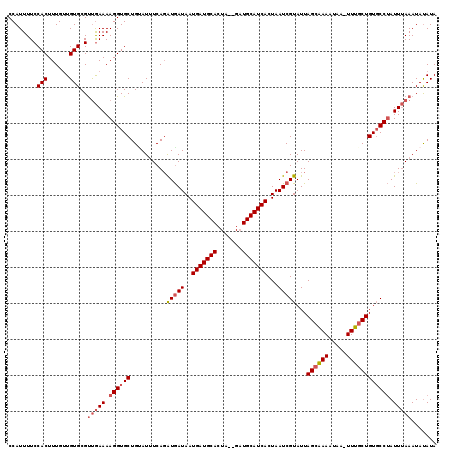
| Location | 9,968,318 – 9,968,434 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9968318 116 - 23771897 UAUAUAUUUAAAUAGGCACAGGAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAGC--UAGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGG ...............((..((.(((-....))).)).....(((.(((((((((..--...))))))))))))............)).(((((((..(((........)))))))).)) ( -20.30) >DroSec_CAF1 21120 118 - 1 UAUAUAUUUAAAUAGGCACAGCAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGG ...............((..((((((-....)))))).....(((.((((((((((.....)))))))))))))............)).(((((((..(((........)))))))).)) ( -26.20) >DroSim_CAF1 28701 118 - 1 UAUAUAUUUAAAUAGGCACAGCAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGG ...............(((.((((((-....)))))).....(((.((((((((((.....)))))))))))))........)))....(((((((..(((........)))))))).)) ( -26.70) >DroYak_CAF1 21143 117 - 1 UAUGUAUUUUAAUGGGAACAGCCAAUGUAUUUGGCUAAUACGAUUAGUGAUGCAUC--UACUGCAUCAUUAUUACAUGACAUACAGCACCUUUUUGGCGCACAUCAAAGUGGAAAAUGG ..(((((....(((.....((((((.....))))))........((((((((((..--...))))))))))...)))...)))))((.((.....)).))................... ( -29.40) >consensus UAUAUAUUUAAAUAGGCACAGCAAA_UUAUUUUGCUAAUACGAUUAGUGAUGCAUC__UAGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGG ...............((..((((((.....)))))).....(((.((((((((((.....)))))))))))))............)).(((((((.((..........)).))))).)) (-21.61 = -21.68 + 0.06)

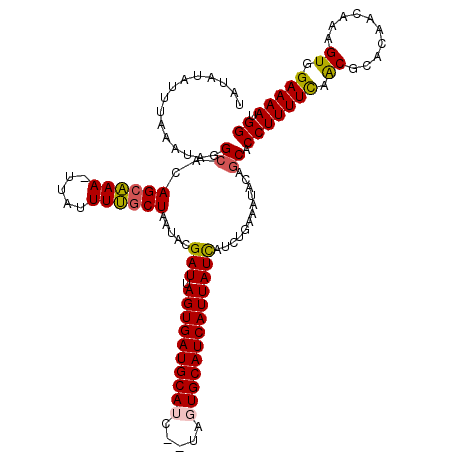

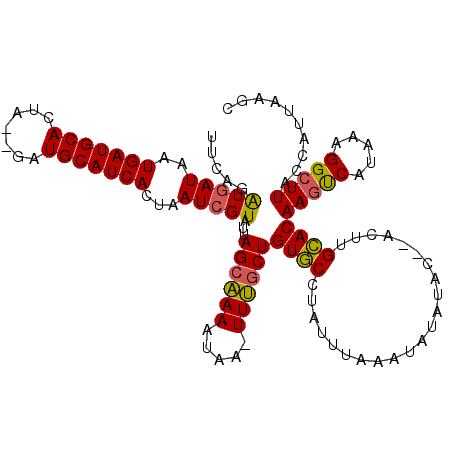
| Location | 9,968,357 – 9,968,468 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -18.30 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9968357 111 + 23771897 UUCAGAUGAUAAUGAUGCACUA--GCUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUCCUGUGCCUAUUUAAAUAUAUAC--ACUUGCACAAGUCAUAAAGGCUUACCAUUAAGC .....(((((..(((((((...--..)))))))...)))))....((.......-......((((................--....))))(((((.....)))))........)) ( -18.25) >DroSec_CAF1 21159 113 + 1 UUCAGAUGAUAAUGAUGCACUGGAGAUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUGCUGUGCCUAUUUAAAUAUAUAC--ACUUGCACAAAUCAUAAAGGCUUACCAUUAAGC .....(((((..((((((((....).)))))))......(((..((((((....-))))))(((..(((......)))..)--)).)))....)))))....(((((....))))) ( -20.90) >DroSim_CAF1 28740 113 + 1 UUCAGAUGAUAAUGAUGCACUAGAGAUGCAUCACUAAUCGUAUUAGCAAAAUAA-UUUGCUGUGCCUAUUUAAAUAUAUAC--ACUUGCACAAGUCAUAAAGGCUUACCAUUAAGC .....(((((..((((((((....).)))))))...)))))...((((((....-))))))((((................--....))))(((((.....))))).......... ( -21.75) >DroYak_CAF1 21182 114 + 1 GUCAUGUAAUAAUGAUGCAGUA--GAUGCAUCACUAAUCGUAUUAGCCAAAUACAUUGGCUGUUCCCAUUAAAAUACAUAUACAGUUGAACAAGCCAUAAAGGCUUACCAUUAGGC ((((((((....(((((((...--..))))))).((((.(...(((((((.....)))))))...).))))...)))))............(((((.....))))).......))) ( -27.70) >consensus UUCAGAUGAUAAUGAUGCACUA__GAUGCAUCACUAAUCGUAUUAGCAAAAUAA_UUUGCUGUGCCUAUUUAAAUAUAUAC__ACUUGCACAAGUCAUAAAGGCUUACCAUUAAGC .....(((((..(((((((.......)))))))...)))))...((((((.....))))))((((......................))))(((((.....))))).......... (-18.30 = -17.93 + -0.37)


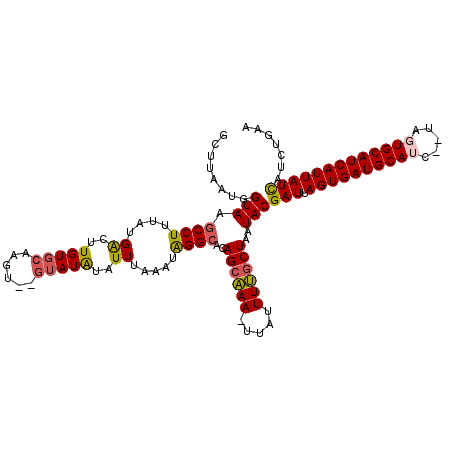
| Location | 9,968,357 – 9,968,468 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9968357 111 - 23771897 GCUUAAUGGUAAGCCUUUAUGACUUGUGCAAGU--GUAUAUAUUUAAAUAGGCACAGGAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAGC--UAGUGCAUCAUUAUCAUCUGAA (((((....)))))........(((((((....--.((......)).....)))))))...-...............(((.(((((((((..--...))))))))))))....... ( -26.10) >DroSec_CAF1 21159 113 - 1 GCUUAAUGGUAAGCCUUUAUGAUUUGUGCAAGU--GUAUAUAUUUAAAUAGGCACAGCAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAA ........(((.((((....(((.(((((....--))))).))).....))))..((((((-....))))))..)))(((.((((((((((.....)))))))))))))....... ( -31.60) >DroSim_CAF1 28740 113 - 1 GCUUAAUGGUAAGCCUUUAUGACUUGUGCAAGU--GUAUAUAUUUAAAUAGGCACAGCAAA-UUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAA ........(((.((((....((..(((((....--)))))..)).....))))..((((((-....))))))..)))(((.((((((((((.....)))))))))))))....... ( -30.60) >DroYak_CAF1 21182 114 - 1 GCCUAAUGGUAAGCCUUUAUGGCUUGUUCAACUGUAUAUGUAUUUUAAUGGGAACAGCCAAUGUAUUUGGCUAAUACGAUUAGUGAUGCAUC--UACUGCAUCAUUAUUACAUGAC (((....)))(((((.....)))))..(((..((((..((((((.....(....)((((((.....))))))))))))..((((((((((..--...)))))))))).))))))). ( -33.10) >consensus GCUUAAUGGUAAGCCUUUAUGACUUGUGCAAGU__GUAUAUAUUUAAAUAGGCACAGCAAA_UUAUUUUGCUAAUACGAUUAGUGAUGCAUC__UAGUGCAUCAUUAUCAUCUGAA ........(((.((((....((..(((((......)))))..)).....))))..((((((.....))))))..)))(((.((((((((((.....)))))))))))))....... (-23.25 = -23.62 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:09 2006