
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,956,063 – 9,956,165 |
| Length | 102 |
| Max. P | 0.850568 |

| Location | 9,956,063 – 9,956,165 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9956063 102 + 23771897 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG---CCCAACAAUUCACAGUCAAA-AUACAUAGGAAGAA-AUUC----AAUGCACAUU (((((....((..(((((....)))))..))(((....)))((((.(((((...---)))))....)))).......-........(((...-.)))----..)))))... ( -32.70) >DroSec_CAF1 9091 101 + 1 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG---C-CAACAAUUCACAGUCAAA-AUACAUGGAAAGAA-AUUC----AAUGCACAUU (((((....((..(((((....)))))..))(((....)))((((.((((....---)-)))....)))).......-..............-....----..)))))... ( -28.80) >DroSim_CAF1 15991 102 + 1 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG---CCCAACAAUUCACAGUCAAA-AUACAUGGAAAGAA-AUUC----AAUGCACAUU (((((....((..(((((....)))))..))(((....)))((((.(((((...---)))))....)))).......-..............-....----..)))))... ( -30.60) >DroEre_CAF1 9098 106 + 1 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG---CCCAGCAAUUCACAGUCAAA-AUACAUAGGACGAA-AUACUGGAAAUUCACAUU (((.((((.((..(((((....)))))..))(((....)))((((.(((((...---)))))....)))).(((...-........)))...-........)))))))... ( -27.10) >DroYak_CAF1 8804 107 + 1 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG---CCCAACAAUUCACAGUCAAA-AUACAUAGAAAGAAAAAACUGAAAACUCUCAUU ......((((...(((((....)))))(((((((....)))((((.(((((...---)))))....)))).......-.......)))).......))))........... ( -24.20) >DroAna_CAF1 10194 91 + 1 GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCACCACCCCACCAUUCACAGUCACAUCUACAGAUACAGA-------------------- ((((.....((..(((((....)))))..))(((....)))((((((((((.......))))........)))))).......).)))...-------------------- ( -26.90) >consensus GUGCAAUUAGAUUUCCGCUAAAGCGGAUUUCGCGAUAUCGCGUGACUUGGGCCG___CCCAACAAUUCACAGUCAAA_AUACAUAGAAAGAA_AUUC____AAUGCACAUU (((......((..(((((....)))))..))(((....)))((((.(((((......)))))....)))).........)))............................. (-22.89 = -23.08 + 0.19)

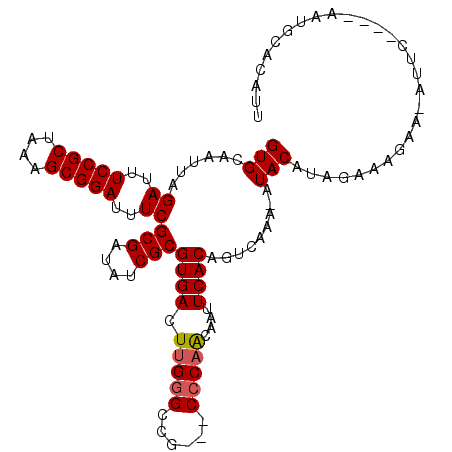

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:03 2006